EagI-HF®
EagI-HF has been reformulated with Recombinant Albumin (rAlbumin) beginning with Lot #10215285. Learn more.
We are excited to announce that all reaction buffers are now BSA-free. NEB began switching our BSA-containing reaction buffers in April 2021 to buffers containing Recombinant Albumin (rAlbumin) for restriction enzymes and some DNA modifying enzymes. Find more details at www.neb.com/BSA-free.

High-Fidelity (HF®) restriction enzymes have the same specificity as native enzymes, but have been engineered for significantly reduced star activity and performance in a single buffer (rCutSmart™ Buffer). All HF-restriction enzymes come with Gel Loading Dye, Purple (6X). Enjoy the enhanced performance and added value of our engineered enzymes at the same price as the native enzyme:
- Engineered for improved performance
- 100% activity in rCutSmart Buffer
- Time-Saver™ qualified for digestion in 5-15 minutes
- Reduced star activity
- Supplied with 1 vial of Gel Loading Dye, Purple (6X)
- Restriction Enzyme Cut Site: C/GGCCG
Featured Video
-
Product Information
High Fidelity (HF) Restriction Enzymes have 100% activity in rCutSmart Buffer; single-buffer simplicity means more straightforward and streamlined sample processing. HF enzymes also exhibit dramatically reduced star activity. HF enzymes are all Time-Saver qualified and can therefore cut substrate DNA in 5-15 with the flexibility to digest overnight without degradation to DNA. Engineered with performance in mind, HF restriction enzymes are fully active under a broader range of conditions, minimizing off-target products, while offering flexibility in experimental design.
NEB extensively performs quality controls on all standard and high-fidelity (HF) restriction enzymes. Examples of nuclease contamination studies for some of our HF restriction enzymes are shown below.
Restriction Enzyme Competitor Study: Nuclease Contamination
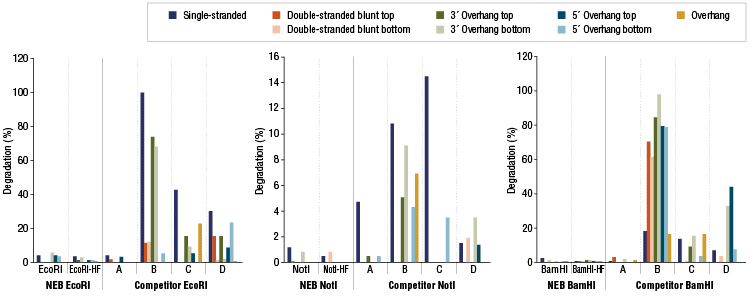
EcoRI, NotI, and BamHI from multiple suppliers were tested in reactions containing a fluorescent labeled single stranded, double stranded blunt, 3’overhang or 5’ overhang containing oligonucleotides. The percent degradation is determined by capillary electrophoresis and peak analysis. The resolution is at the single nucleotide level.Product Source
An E. coli strain that carries the cloned and modified EagI gene from Enterobacter agglomerans (R. Morgan)- This product is related to the following categories:
- Restriction Endonucleases C G Products,
- High-Fidelity (HF®) Restriction Endonucleases Products,
- Time-Saver Qualified Restriction Enzymes Products
- This product can be used in the following applications:
- Fast Cloning: Accelerate your cloning workflows with reagents from NEB,
- Restriction Enzyme Digestion
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
-

Reduce Star Activity with High-Fidelity Restriction Enzymes
-

TIME-SAVER™ Protocol for Restriction Enzyme Digests
-

NEB® TV Ep. 15 – Applications of Restriction Enzymes
-

Restriction Enzyme Digest Protocol: Cutting Close to DNA End
-

Restriction Enzyme Digestion Problem: DNA Smear on Agarose Gel
-
Why is My Restriction Enzyme Not Cutting DNA?
-

Restriction Enzyme Digest Problem: Too Many DNA Bands
-
Double Digestion with NEBcloner
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.