BsaI
This product was discontinued on 12/31/2020 and replaced with the High Fidelity version, BsaI-HFv2 (NEB #R3733).
- Catalog # R0535 was discontinued on December 31, 2020

Attention Golden Gate Assembly users: BsaI-HFv2 (NEB #R3733) has been optimized for Golden Gate Assembly*. BsaI-HFv2 also works well for any protocol requiring DNA cutting by BsaI. This is the recommended enzyme for any purpose requiring digestion at the recognition sequence ...5′-GGTCTC(N1)/(N5)-3′...
*The requirements for digestion of DNA during Golden Gate Assembly are more demanding than what is encountered in traditional DNA cloning. For Golden Gate, the enzyme must function well in a buffer that might not be the same buffer as that normally provided for the restriction enzyme, and maintain activity for an extended time at elevated temperatures in a dynamic cutting/re-ligating reaction with competition for substrate binding between the endonuclease and ligase. Extensive testing has demonstrated superior performance of BsaI-HFv2 compared to both BsaI (NEB #R0535) and BsaI-HF (NEB #R3535) in this challenging Golden Gate assembly context where restriction enzyme efficiency and fidelity are critically important.
- High Fidelity (HF®) version available (NEB #R3733) supplied with CutSmart® Buffer
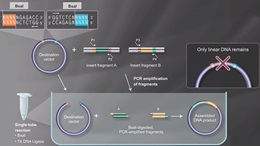
- Used in Golden Gate Assembly
- Type IIS restriction enzymes recognize asymmetric DNA sequences and cleave outside of their recognition sequence
- 100% activity in CutSmart Buffer (over 210 enzymes are available in the same buffer) simplifying double digests
- Supplied with 1 vial of Gel Loading Dye, Purple (6X)
Featured Video
-
Product Information
BsaI has a High Fidelity version BsaI-HF®v2 (NEB #R3733).
High Fidelity (HF) Restriction Enzymes have 100% activity in CutSmart Buffer; single-buffer simplicity means more straightforward and streamlined sample processing. HF enzymes also exhibit dramatically reduced star activity. HF enzymes are all Time-Saver qualified and can therefore cut substrate DNA in 5-15 minutes with the flexibility to digest overnight without degradation to DNA. Engineered with performance in mind, HF restriction enzymes are fully active under a broader range of conditions, minimizing off-target products, while offering flexibility in experimental design.Product Source
An E. coli strain that carries the cloned BsaI gene from Bacillus stearothermophilus 6-55 (Z. Chen)- This product is related to the following categories:
- Discontinued (<3 years)
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
-
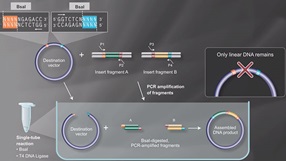
Golden Gate Assembly Workflow
-
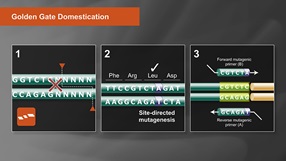
Golden Gate Assembly Domestication Tutorial
-

Golden Gate Assembly Tool Tutorial
-

Restriction Enzymes in Golden Gate Assembly
-

Reduce Star Activity with High-Fidelity Restriction Enzymes
-

Standard Protocol for Restriction Enzyme Digests
-

NEB® TV Ep. 15 – Applications of Restriction Enzymes
-

Restriction Enzyme Digest Protocol: Cutting Close to DNA End
-

Restriction Enzyme Digestion Problem: DNA Smear on Agarose Gel
-
Why is My Restriction Enzyme Not Cutting DNA?
-

Restriction Enzyme Digest Problem: Too Many DNA Bands
-
Double Digestion with NEBcloner
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.