Gibson Assembly® Cloning Kit
Gibson Assembly Master Mix (NEB #M5510) has been reformulated with components containing Recombinant Albumin (rAlbumin) beginning with Lot #10229799.
Have you tried NEBuilder HiFi DNA Assembly? NEBuilder HiFi offers several advantages over NEB Gibson Assembly. For more information, visit NEBuilderHiFi.com.
Gibson Assembly® allows for successful assembly of multiple DNA fragments, regardless of fragment length or end compatibility.
- Assembly and transformation in just under two hours
- Flexible sequence design (scar-less cloning)
- No PCR clean-up step required
- High transformation efficiencies for inserts up to 20 kb
- Easily adapted for multiple DNA manipulations, including site-directed mutagenesis
- Includes competent cells
Featured Video
-
Product Information

Gibson Assembly was developed by Dr. Daniel Gibson and his colleagues at the J. Craig Venter Institute and licensed to NEB by Synthetic Genomics, Inc. It allows for successful assembly of multiple DNA fragments, regardless of fragment length or end compatibility. It has been rapidly adopted by the synthetic biology community due to its ease-of-use, flexibility and suitability for large DNA constructs.
Gibson Assembly efficiently joins multiple overlapping DNA fragments in a single-tube isothermal reaction (1,2). The Gibson Assembly Master Mix includes three different enzymatic activities that perform in a single buffer:
- The exonuclease creates single-stranded 3´ overhangs that facilitate the annealing of fragments that share complementarity at one end (overlap region).
- The proprietary DNA polymerase fills in gaps within each annealed fragment.
- The DNA ligase seals nicks in the assembled DNA.
The end result is a double-stranded fully sealed DNA molecule that can serve as template for PCR, RCA or a variety of other molecular biology applications, including direct transformation. The method has been successfully used by Gibson’s group and others to assemble oligonucleotides, DNA with varied overlaps (15–80 bp) and fragments hundreds of kilobases long (1–2).
To help select the best DNA assembly method for your needs, please use our Synthetic Biology/DNA Assembly Selection Chart.
For help designing primers, please view our primer design video.
Overview of the Gibson Assembly Cloning Method
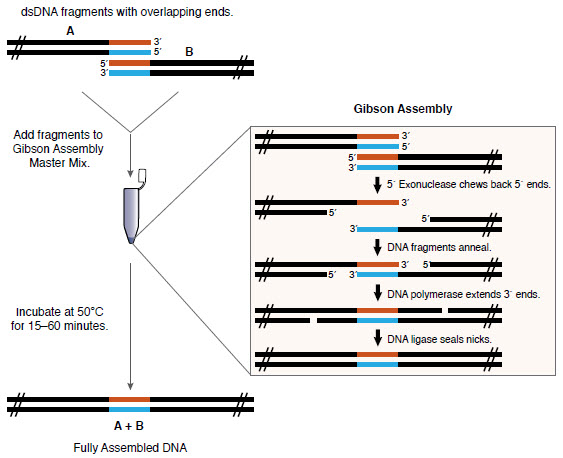
10 μl of 2X Gibson Assembly Master Mix was incubated with 6 fragments (5 fragments of 400 bp and one of 2,780 bp, with 40 bp overlap, 0.05 pmol each) in a final volume of 20 μl at 50°C for 60 minutes. NEB 5-alpha Competent E. coli (NEB #C2987) were transformed with 2 μl of the master mix/fragment mixture using the transformation protocol on page 12. Greater than 100 white colonies were observed when 1/10 of the outgrowth was spread on an ampicillin plate with IPTG/Xgal and incubated overnight.
Specification:
Overview of Gibson Assembly Cloning Kit Protocol:
- Design primers to amplify fragments (and/or vector) with appropriate overlaps
- PCR amplify fragments using a high-fidelity DNA polymerase.
- Prepare linearized vector by PCR amplification using a high-fidelity DNA polymerase or by restriction digestion.
- Confirm and determine concentration of fragments and linearized vector using agarose gel electrophoresis, a NanoDrop™ instrument or other method.
- Add fragments and linearized vector to Gibson Assembly Master Mix and incubate at 50°C for 15 minutes to 1 hour, depending on number of fragments being assembled.
- Transform into NEB 5-alpha Competent E. coli (provided) or use directly in other applications.
- This product is related to the following categories:
- DNA Assembly, Cloning and Mutagenesis Kits Products
- This product can be used in the following applications:
- Gibson Assembly®
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
-
Introduction to Gibson Assembly®
-
Primer Design and Fragment Assembly Using NEBuilder HiFi DNA Assembly® or Gibson Assembly®
-

Gibson Assembly Workflow
-

NEBUILDER® Assembly Tool 2.0 What’s New?
-

NEBUILDER® Assembly Tool 2.0 Fragments Amplified by PCR
-

NEBUILDER® Assembly Tool 2.0 Restriction Enzyme Digest
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.