Q5® Hot Start High-Fidelity DNA Polymerase
Fidelity at its finest – for over 10 years
This is product is available in a glycerol-free format. Contact us for more information.
Q5® High-Fidelity DNA Polymerase sets a new standard for both fidelity and robust performance. With the highest fidelity amplification available (~280 times higher than Taq), Q5 results in ultra-low error rates. Q5 is composed of a novel polymerase that is fused to the processivity-enhancing Sso7d DNA binding domain, improving speed, fidelity and reliability of performance.
Working with uracil-containing DNA templates or using dUTP? Learn about Q5U Hot Start High-Fidelity DNA Polymerase (NEB #M0515).
- Highest fidelity amplification (~280X higher than Taq)
- Ultra-low error rates
- Superior performance for a broad range of amplicons (from high AT to high GC)
- Also available: Q5 standalone polymerase (NEB #M0491) and master mix formats (NEB #M0492, M0494)
Featured Video
-
Product Information
Q5® Hot Start High-Fidelity DNA Polymerase is a high-fidelity, thermostable, hot start DNA polymerase with 3´→ 5´ exonuclease activity, fused to a processivity-enhancing Sso7d domain to support robust DNA amplification. The addition of an aptamer-based inhibitor allows room temperature reaction setup. With an error rate ~280-fold lower than that of Taq DNA Polymerase, Q5 Hot Start High-Fidelity DNA Polymerase is ideal for cloning and can be used for long or difficult amplicons. Q5 Hot Start High-Fidelity DNA Polymerase is supplied with an optimized buffer system that allows robust amplification regardless of GC content. The 5X Q5 Reaction Buffer contains 2 mM Mg++ at final (1X) reaction concentrations and is recommended for most routine applications. For GC-rich targets (≥ 65% GC), amplification can be improved by the addition of the 5X Q5 High GC Enhancer. Q5 Hot Start High-Fidelity DNA Polymerase is unlike typical, lower fidelity PCR enzymes. To determine the optimal annealing temperatures for a given set of primers, use of the NEB Tm Calculator is highly recommended.
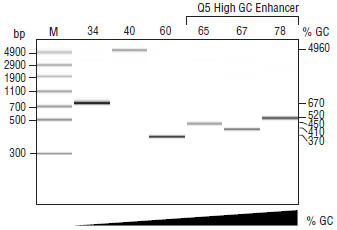
Amplification of a variety of human genomic amplicons from low to high GC content using Q5 Hot Start High-Fidelity DNA Polymerase. All reactions were set up at room temperature, conducted using 30 cycles of amplification and visualized by microfluidic LabChip® analysis. 
Product Source
An E. coli strain that carries the Q5 High-Fidelity DNA Polymerase gene.- This product is related to the following categories:
- Q5® High-Fidelity DNA Polymerases Products
- This product can be used in the following applications:
- Hot Start PCR,
- Long Range PCR,
- Fast PCR,
- Genome Editing Applications,
- High-Fidelity PCR,
- Multiplex PCR,
- Specialty PCR,
- Routine PCR,
- DNA Amplification, PCR & qPCR,
- PCR, Fast Cloning: Accelerate your cloning workflows with reagents from NEB
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
-

Important Tips for Q5® Polymerase
-

Why Choose Q5® High-fidelity Polymerase?
-

Behind the paper: Examining Sources of Error in PCR by Single-Molecule Sequencing
-

Why is Tm Important in Primer Design?
-

Tips for Amplifying Large Amplicons
-
How to Amplify GC-rich DNA
-

5 Tips for Setting Up Your PCR
-
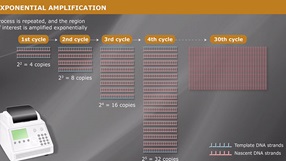
Overview of PCR
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.