Genome Editing
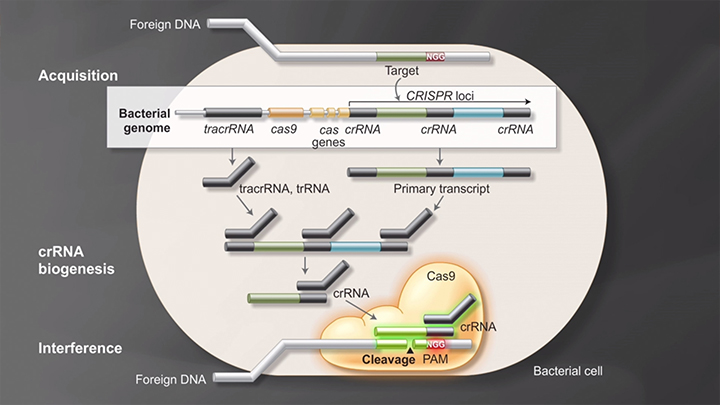
Cas9 in vivo: Bacterial adaptive immunity
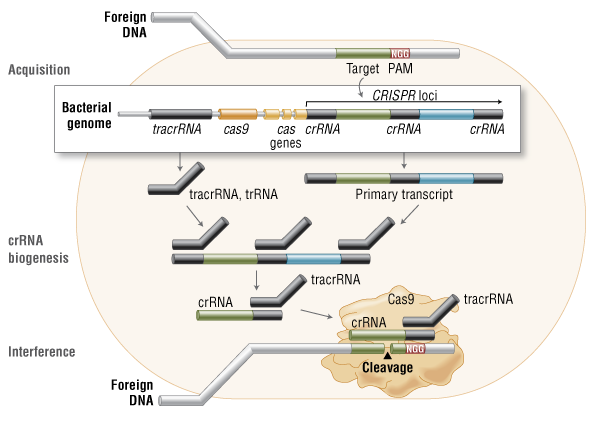
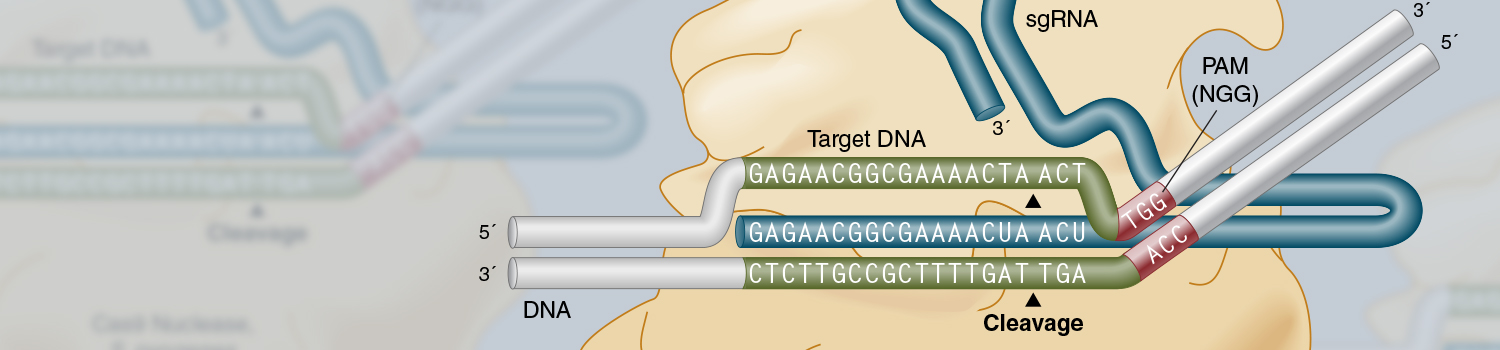
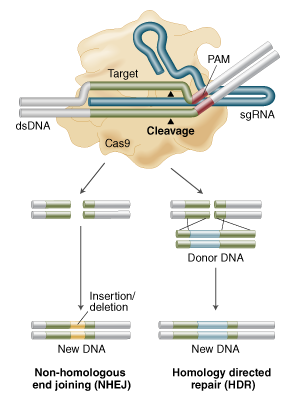
Genome engineering with Cas9 Nuclease
Choose Type:
- What reaction conditions were used to define Authenticase™?
- How does Authenticase™ improve the quality and fidelity of PCR gene assembly?
- How do I convert my gene of interest into oligonucleotides?
- Why do you recommend setting up two tubes for the PCR reaction containing different amounts of Authenticase™-treated samples as templates?
- Can I use Authenticase™ for genome editing applications?
- Does Authenticase™ recognize single base pair mismatches or indels (insertions/deletions)?
- What common additives inhibit Authenticase™?
- What PCR reagents are recommended for DNA amplification in genome editing (CRISPR/Cas9, TALEN, ZFN) mismatch detection assays?
- Can I use Authenticase™ genome editing (CRISPR/Cas9, TALEN, ZFN) mismatch detection assays with unpurified PCR products?
- What size PCR amplicon should I design to analyze the genomic editing efficiency?
- If my PCR reaction yield is low, can I add more than 5 µl of the PCR reaction to the digestion reaction?
- Why do I see an extra band when I run the undigested heteroduplex on a Bioanalyzer or agarose gel?
- What are the differences between Mismatch Endonuclease I (NEB #M0678) and Authenticase (NEB #M0689)?
- PCR Using Q5® Hot Start High-Fidelity DNA Polymerase (M0493)
- In vitro digestion of DNA with Cas9 Nuclease, S. pyogenes (M0386)
- Determining Genome Targeting Efficiency using T7 Endonuclease I
- NEBuilder HiFi DNA Assembly Reaction Protocol
- NEBuilder® HiFi DNA Assembly Electrocompetent Transformation Protocol
- NEBuilder® HiFi DNA Assembly Chemical Transformation Protocol (E2621, E5520, E2623)
- Using recombinant Cas9 nuclease to assess locus modification in genome editing experiments (#M0386)
- sgRNA Synthesis Using the HiScribe™ Quick T7 High Yield RNA Synthesis Kit (NEB #E2050)
- T7 Endonuclease I-based Mutation Detection with the EnGen® Mutation Detection Kit (NEB #E3321)
- EnGen® sgRNA Synthesis Kit, S. pyogenes Protocol (E3322)
- Transfection of Cas9 RNP (ribonucleoprotein) into adherent cells using the Lipofectamine® RNAiMAX
- Error Correction During Gene Synthesis (NEB #M0689)
- Supplemental Protocol 1: Generation of DNA fragments by PCR assembly of pooled oligos (NEB #M0689)
- Mismatch Detection Assay (NEB #M0689)
- Supplemental Protocol 2: Using colony PCR to identify positive clones (NEB #M0689)
- Transfection of EnGen® Spy Cas9 HF1 (NEB #M0667) into adherent cells using the Lipofectamine® RNAiMAX System
- Rapid Analysis of Genome Editing Efficiency using PCR Amplicons processed by the Exo-CIP Rapid PCR Cleanup Kit followed by Sanger sequencing
- A faster workflow for the assessment of genomic loci in mice using a novel HMW DNA extraction technology upstream of Cas9 targeted sequencing
- Determining Efficiency of On-Target CRISPR Cas9 Genome Editing Using the NEB® EnGen® Mutation Detection Kit on LabChip Gel Xpress
- Bridging dsDNA with a ssDNA Oligo and NEBuilder® HiFi DNA Assembly to create an sgRNA-Cas9 Expression Vector
- Protocol for using Recombinant Cas9 Nuclease to Assess Locus Modification in Genome Editing Experiments
- Construction of an sgRNA-Cas9 expression vector via single-stranded DNA oligo bridging of double-stranded DNA fragments
-
CRISPR/Cas9 & Targeted Genome Editing: New Era in Molecular Biology
Understand the history, importance and future of CRISPR/Cas9 and target genome editing
- Genome Editing Brochure
- Kinetic Comparison of Cas9 Homologs Recognizing Diverse PAM Sequences (2018)
Feature Articles
Brochures
Posters
- Zheng Hu, Wencheng Ding, Da Zhu, Lan Yu, Xiaohui Jiang, Xiaoli Wang, Changlin Zhang, Liming Wang, Teng Ji, Dan Liu, Dan He, Xi Xia, Tao Zhu, Juncheng Wei, Peng Wu, Changyu Wang, Ling Xi, Qinglei Gao, Gang Chen, Rong Liu, Kezhen Li, Shuang Li, Shixuan Wang, Jianfeng Zhou, Ding Ma, Hui Wang (2015) TALEN-mediated targeting of HPV oncogenes ameliorates HPV-related cervical malignancy. J Clin Invest; 125, 425-36. PubMedID: 25500889, DOI: 10.1172/JCI78206
- A-Bk Dad, S Ramakrishna, M Song, H Kim (2014) Enhanced gene disruption by programmable nucleases delivered by a minicircle vector. Gene Ther; PubMedID: 25142139, DOI: 10.1038/gt.2014.76
Online Resources
Plasmid Repositories:Addgene
CRISPR-gRNA Design Tools:
Desktop Genetics
CRISPR Design
ZiFiT
CHOP CHOP
Online Forums:
Genome Engineering using CRISPR/Cas Systems
Organism-specific Resources:
Cas9-triggered homologous recombination
Drosophila RNAi Screening Center at Harvard Medical School
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.