T7 Endonuclease I-based Mutation Detection with the EnGen® Mutation Detection Kit (NEB #E3321)
New users are encouraged to perform PCR and T7 Endonuclease I digestion using the included control template and primer mix. For each amplicon we recommend setting up three PCR reactions using the following templates:
a. gDNA from targeted cells (e.g., Cas9, TALEN or ZFN transfected cells)
b. gDNA from negative control cells (e.g., non-specific DNA transfected cells)
c. Water (i.e., no template control)
Amplification reactions for experimental samples
-
Thaw the kit components, mix and pulse-spin in microfuge each
component prior to use.
- Set up a 25 μl PCR reaction using up to 500 ng of genomic DNA as a
template.
Assemble the following reaction at room temperature:
* To use cell lysate directly in PCR, lyse cells in QuickExtract or DNAzol Direct using 50 μl cells in each well of a 96-well plate (~40,000 cells) according to the manufacturers’ recommendation. Dilute the lysate 1:5 in TE and use 2.5 μl of the diluted lysate.REAGENT 25 μl RXN FINAL CONCENTRATION Q5 Hot Start High-Fidelity 2X Master Mix 12.5 μl 1X 10 μM Forward Primer 1.25 μl 0.5 μM 10 μM Reverse Primer 1.25 μl 0.5 μM Template DNA variable 0.5–500 ng genomic DNA* Nuclease-free water to 25 μl
- Gently mix the reaction. Collect all the liquid to the bottom of the tube
with a brief spin. Transfer the tubes to a PCR machine and begin thermocycling
using the following conditions:
* Please visit Tmcalculator.neb.com to determine correct annealing temperature.CYCLE STEP TEMP TIME CYCLES; Initial Denaturation 98°C 30 seconds 1 Denaturation
Annealing
Extension98°C
50–72°C*
72°C5 seconds
10 seconds
20 seconds35 Final Extension 72°C 2 minutes
1 Hold 4–10°C
Control reaction using included Control Template and Primer Mix.
-
Set up a 25 μl PCR reaction as follows:
REAGENT 25 μl RXN FINAL CONCENTRATION Q5 Hot Start High-Fidelity 2X Master Mix 12.5 μl 1X Control Template and Primer Mix 2.5 μl 0.5 ng plasmid
and 0.5 μM each primerNuclease-free water 10 μl
- Gently mix the reaction. Collect all the liquid to the bottom of the tube
with a brief spin. Transfer the tubes to a PCR machine and begin thermocycling
using the following conditions:
CYCLE STEP TEMP TIME CYCLES; Initial Denaturation 98°C 30 seconds 1 Denaturation
Annealing
Extension98°C
65°C
72°C5 seconds
10 seconds
20 seconds35 Final Extension 72°C 2 minutes
1 Hold 4–10°C
- Analyze a small amount of the PCR product on an agarose gel to verify amplification of a single product of the correct size. A DNA marker should also be run to help estimate amplicon concentration. The product of the control PCR reaction is ~600 bp.
Heteroduplex formation:
The products of the PCR reaction must be denatured and annealed in order to allow formation of heteroduplex between PCR products with and without mutations. T7 Endonuclease I digestion has been optimized for use with 5 μl of the PCR reaction, containing up to 250 ng of amplified DNA.The following protocol applies to both experimental and control reactions:
- Assemble the reaction as follows:
REAGENT 19 μl ANNEALING REACTION PCR Reaction 5 μl 10X NEBuffer 2 2 μl Nuclease-free water 12 μl
- Denature and then anneal the products in a thermocycler using the following
program*:
* Alternatively, if a thermocycler is not available with these ramp speeds, the sample can be heated to 95ºC for 10 minutes and then allowed to cool slowly to room temperature.CYCLE STEP
TEMP
RAMP RATE
TIME
Initial Denaturation
95°C
5 minutes
Annealing
95–85°C
85–25°C-2°C/second
-0.1°C/second
Hold
4°C
- Proceed to heteroduplex digestion.
Heteroduplex digestion:
The digestion reaction conditions have been optimized for 5 μl of the unpurified PCR reaction containing up to 250 ng of amplified DNA. Increased amounts of PCR reaction and/or DNA may lead to inaccurate estimates of editing efficiencies.- Set up each reaction as follows:
REAGENT 20 μl T7E1 REACTION Annealed PCR Product 19 μl EnGen T7 Endonuclease I 1 μl
- Mix well and briefly spin. Incubate each reaction at 37°C for 15 minutes.
- Following digestion, add 1 μl of Proteinase K and mix well.
- Incubate for 5 minutes at 37°C to inactivate the T7 Endonuclease I.
- Proceed with fragment analysis or store at –20°C until ready.
Optional: If needed, reactions can be purified prior to fragment analysis. For this we recommend the Monarch® PCR & DNA Cleanup Kit (5 μg) (NEB #T1030) .
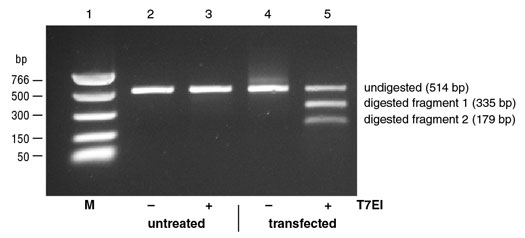
Figure 2: Example of Mutation Detection on targeted 293 cells.
Analysis of DNA Fragments:
1. Gel Analysis
Add 4 μl of Gel Loading Dye, Purple (6X), no SDS (NEB #B7025 ) to the reaction and run on a 2% agarose gel stained with ethidium bromide. Run the included DNA ladder or an appropriate DNA size marker along side the sample for reference.
Alternatively, samples can be analyzed using a fragment analyzer (e.g.,Agilent Bioanalyzer or Advanced Analytical Technologies, Inc (AATI) Fragment Analyzer). For the Agilent Bioanalyzer, 1 μl of the reaction will not interfere when using the standard sensitivity Agilent DNA analysis kits. For the AATI Fragment Analyzer, 2 μl of the reaction can be used with the Standard Sensitivity NGS Fragment Analysis Kit (AATI Cat# DNF-473).
2. Calculate the estimated % modification using the following formula:
% Modification = 100 x [1-(1-fraction cleaved)1/2]
When calculating % modification for reactions with the control template
where the starting material is known, the equation (100 x fraction
cleaved) can be used.
Where fraction cleaved = concentration of digested products/
(concentration of digested products + concentration of undigested band)

