Shrimp Alkaline Phosphatase (rSAP)
We are excited to announce that all reaction buffers are now BSA-free. NEB began switching our BSA-containing reaction buffers in April 2021 to buffers containing Recombinant Albumin (rAlbumin) for restriction enzymes and some DNA modifying enzymes. Find more details at www.neb.com/BSA-free.
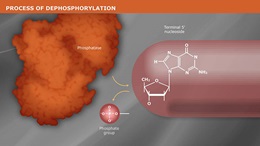
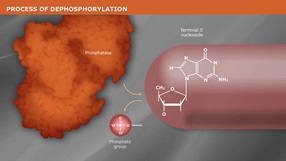
Shrimp Alkaline Phosphatase (rSAP) is a heat labile alkaline phosphatase purified from a recombinant source.
- Rapid and irreversible heat inactivation eliminates unwanted activity
- Improved storage stability versus native enzyme
- Faster reaction setup (no supplemental additives like zinc required)
- Flexible reaction conditions (active in any restriction enzyme buffer, no clean-up required)
- Less enzyme required (high specific activity), resulting in a lower cost per reaction
- No need for multiple phosphatases (rSAP removes 5´- and 3´- phosphates from DNA, RNA and dNTPs )
- Active on unincorporated dNTPs in PCR products - improves DNA sequencing and SNP analysis
- Recombinant for purity, consistency and value
Featured Video
-
Product Information
Shrimp Alkaline Phosphatase (rSAP) is a heat labile alkaline phosphatase purified from a recombinant source. rSAP is identical to the native enzyme and contains no affinity tags or other modifications. rSAP nonspecifically catalyzes the dephosphorylation of 5´ and 3´ ends of DNA and RNA phosphomonoesters. Also, rSAP hydrolyses ribo-, as well as deoxyribonucleoside triphosphates (NTPs and dNTPs). rSAP is useful in many molecular biology applications such as the removal of phosphorylated ends of DNA and RNA for subsequent use in cloning or end-labeling of probes. In cloning, dephosphorylation prevents religation of linearized plasmid DNA. The enzyme acts on 5´ protruding, 5´ recessed and blunt ends. rSAP may also be used to degrade unincorporated dNTPs in PCR reactions to prepare templates for DNA sequencing or SNP analysis. rSAP is completely and irreversibly inactivated by heating at 65°C for 5 minutes, thereby making removal of rSAP prior to ligation or end-labeling unnecessary.
Figure 1: rSAP heat inactivation at 65°C
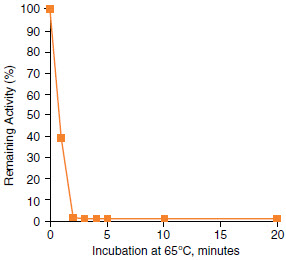
1 unit of rSAP was incubated under recommended reaction conditions, including DNA, for 30 minutes and then heated at 65°C. Remaining phosphatase activity was measured by PNPP assay.
Product Source
A Pichia pastoris clone that carries the shrimp alkaline phosphatase gene from Northern shrimp Pandalus borealis (1,2).- This product is related to the following categories:
- Phosphatases Products
- This product can be used in the following applications:
- Dephosphorylation
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.