T4 DNA Polymerase
We are excited to announce that all reaction buffers are now BSA-free. NEB began switching our BSA-containing reaction buffers in April 2021 to buffers containing Recombinant Albumin (rAlbumin) for restriction enzymes and some DNA modifying enzymes. Find more details at www.neb.com/BSA-free.
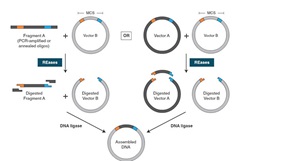
T4 DNA Polymerase catalyzes the synthesis of DNA in the 5´→ 3´ direction and requires the presence of template and primer.
- Gap filling (no strand displacement activity)
- Removal of 3’ overhangs or fill-in of 5’ overhangs to form blunt ends
- Lacks 5’ —> 3’ exonuclease activity
- Probe labeling using replacement synthesis
- Singe-strand deletion subcloning
Featured Video
-
Product Information
T4 DNA Polymerase catalyzes the synthesis of DNA in the 5´→ 3´ direction and requires the presence of template and primer. This enzyme has a 3´→ 5´ exonuclease activity which is much more active than that found in DNA Polymerase I (E. coli). Unlike E. coli DNA Polymerase I, T4 DNA Polymerase does not have a 5´→ 3´ exonuclease function.
Product Source
Purified from a strain of E. coli that carries the T4 DNA Polymerase gene.- This product is related to the following categories:
- DNA Manipulation Products
- This product can be used in the following applications:
- Blunting,
- Polymerases for DNA Manipulation,
- PCR
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.