DNA Blunting Tutorial

Script
Blunting a fragment of DNA involves the removal or fill-in of any unpaired, overhanging bases. This process is often used to prepare fragments for ligation with other blunt-ended DNA, or fragments with non-compatible ends.
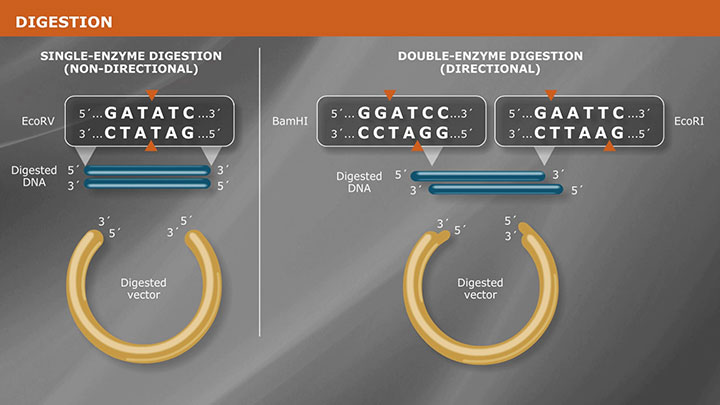
When planning to blunt the ends of your DNA fragment, you’ll need to determine what types of ends your DNA fragment has – a 5´ overhang, a 3´ overhang, or blunt. All three types of ends are possible if your fragments were generated by restriction enzyme digestion.
If your DNA fragment has blunt ends, you can sit back and relax while we demonstrate how to determine if an overhang is a 3´ overhang or a 5´ overhang.
The first step is simply to write out the digested sequence and pattern…then, label the 5´ and 3´ ends of both strands of DNA. Quite simply, the orientation of the protruding end will tell you what type of fragment end you’re dealing with.
Polymerases such as T4 DNA Polymerase and DNA Polymerase I, Large (Klenow) Fragment can blunt an end by either using a polymerase activity to fill in a 5´ overhang in the 5´ to 3´ direction…or, they can blunt a 3´ overhang by degrading the overhang in the 3´ to 5´ direction using an exonuclease activity.
If you have specific requirements about maintaining sequence at the blunting site, and you need to digest the 5´ overhang, you can use Mung Bean Nuclease to degrade in a 5´ to 3´ direction. We only recommend the use of this enzyme in this scenario.
With practice, identifying overhang orientation will get easier, and you won’t need to write out the digested sequence every time… although, it is a good habit to develop.
Use NEB’s online tool, NEBcloner , to find products and protocols for each step in your cloning workflow, including blunting. Visit CloneWithNEB.com for more information, including troubleshooting, technical tips and the full list of products available.
Related Videos
-
Overview of Traditional Cloning -

NEB® Restriction Enzyme Double Digest Protocol

