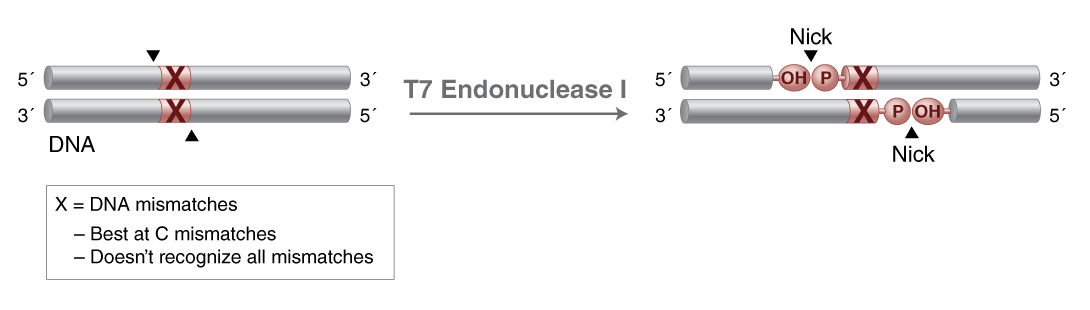
T7 Endonuclease I
Authenticase® (NEB #M0689) now available for improved genome editing mutation detection.
- DNA Endonuclease
- Catalyzes the cleavage of DNA mismatches and non-β DNA structures including Holliday junctions and cruciform leaving 3'-OH and 5'-phosphate
- Best at C mismatches and does not recognize all DNA mismatches
- To a lesser extent cleaves across a nick in dsDNA
- Key enzyme for use in genome editing mutation detection workflows
- Also available in convenient kit format: EnGen Mutation Detection Kit (NEB #E3321)
- Need help finding the right exonuclease for your experiments? Try Exo Selector
Featured Video
-
Product Information
T7 Endonuclease I recognizes and cleaves non-perfectly matched DNA, cruciform DNA structures, Holliday structures or junctions, heteroduplex DNA and more slowly, nicked double-stranded DNA. The cleavage site is at the first, second or third phosphodiester bond that is 5´ to the mismatch. The protein is the product of T7 gene 3.
Highlights
- Isolated from a recombinant source
- Supplied with 10X Reaction Buffer
- Recognizes non-perfectly matched DNA
Product Source
An E. coli strain that carries a fusion of maltose binding protein and T7 Endonuclease I (T7 Endo I).- This product is related to the following categories:
- Mutation Detection Reagents and Kits Products,
- Genome Editing Products,
- Exonucleases and Non-specific Endonucleases Products, DNA Repair Enzymes and Structure-specific Endonucleases Products
- This product can be used in the following applications:
- Whole Genome Amplification & Multiple Displacement Amplification,
- Genome Editing Applications
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.