9°N™ DNA Ligase
9°N DNA Ligase has been reformulated with Recombinant Albumin (rAlbumin) beginning with Lot #10230152.
9°N DNA Ligase will ligate this substrate:
Nicked DNA/RNA
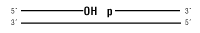
- Can be used to ligate nicks in DNA while incubating at high temperatures
- Extremely thermostable and can withstand PCR conditions
- Will not ligate short 4 base overlaps (typical of restriction enzyme digests), but efficiently ligates 12 base pair overlaps
- Isolated from a recombinant source
- Not sure which ligase to choose? Refer to our DNA and RNA Ligase Properties Chart or DNA Ligase Selection Chart
Featured Video
-
Product Information
9° N™ DNA Ligase is a thermostable ligase that catalyzes the formation of a phosphodiester bond between the 5´-phosphate and the 3´-hydroxyl of two adjacent DNA strands that are hybridized and accurately paired, with no gap, to a complementary DNA strand. 9° N DNA Ligase uses ATP as a cofactor and it is active at elevated temperatures (45° C – 70° C). Product Source
Purified from an E. coli strain containing the cloned ligase gene from the extremely thermophilic marine archaea Thermococcus sp.(strain 9°N). The archaea was isolated from a submarine thermal vent, at a depth of 2,500 meters, 9° north of the equator at the East Pacific Rise (1).- This product is related to the following categories:
- DNA Ligases Products
- This product can be used in the following applications:
- Cloning Ligation
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.