T4 Polynucleotide Kinase
- 5' phosphorylation of DNA/RNA for subsequent ligation
- End labeling DNA or RNA for probes and DNA sequencing
- Removal of 3' phosphoryl groups
Featured Video
-
Product Information
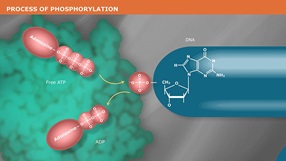
Catalyzes the transfer and exchange of Pi from the γ position of ATP to the 5´ -hydroxyl terminus of polynucleotides (double-and single-stranded DNA and RNA) and nucleoside 3´-monophosphates. Polynucleotide Kinase also catalyzes the removal of 3´-phosphoryl groups from 3´-phosphoryl polynucleotides, deoxynucleoside 3´-monophosphates and deoxynucleoside 3´-diphosphates (1). Product Source
A E. coli strain that carries the cloned T4 Polynucleotide Kinase gene. It is purified by a modification of the method of Richardson (1).- This product is related to the following categories:
- DNA Labeling Products,
- Kinases Products
- This product can be used in the following applications:
- Phosphorylation (Kinase)
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.