Restriction Enzymes for Epigenetic Analysis
Choose Type:
- Optimizing Restriction Endonuclease Reactions
- Protocol for Glucosylation and digestion of Genomic DNA using AbaSI (#R0665)
- Protocol for Direct Digestion of gDNA during droplet digital PCR (ddPCR)
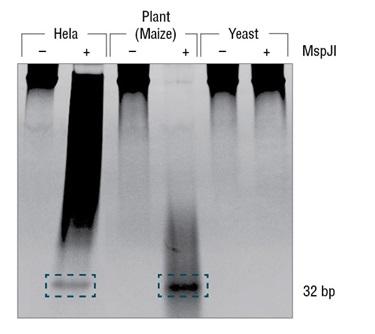
- Protocol for generating 32 bp fragments from modified CpG sites in genomic DNA using MspJI, FspEI or LpnPI
- Double Digest Protocol with Standard Restriction Enzymes
-
Epigenetics - Expanding on Genomic Foundations
-
Restriction Endonucleases: Molecular Cloning and Beyond
- Epigenetics Brochure
- Dam-Dcm and CpG Methylation
- Frequencies of Restriction Sites
- EpiMark® Methylated DNA Enrichment Kit Troubleshooting Guide
- Alteration of Apparent Recognition Specificities Using Methylases
- Dam and Dcm Methylases of E. coli
- Restriction Enzyme Tips
- Restriction of Foreign DNA by E. coli K-12
- Site Preferences
Feature Articles
Brochures
Selection Tools
Troubleshooting Guides
Usage Guidelines
- Mooijman D, Dey S S, Boisset JC, Crosetto N, van Oudenaarden A (2016) Single-cell 5hmC sequencing reveals chromosome-wide cell-to-cell variability and enables lineage reconstruction Nat Biotechnol; 34, 852-857. PubMedID: 27347753, DOI: 10.1038/nbt.3598
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.