Interactive Tutorial Explaining the Phenomenon of Epigenetics at a Molecular Level
Script
Epigenetic regulation encompasses a number of different modifications to the chromatin. These include methylation of the DNA on cytosine bases, a modification that can further be oxidized, as well as modification to the histone tails that emanate from the core of the nucleosome. The tails of the core histones labeled here can be altered with distinct chemical modifications.
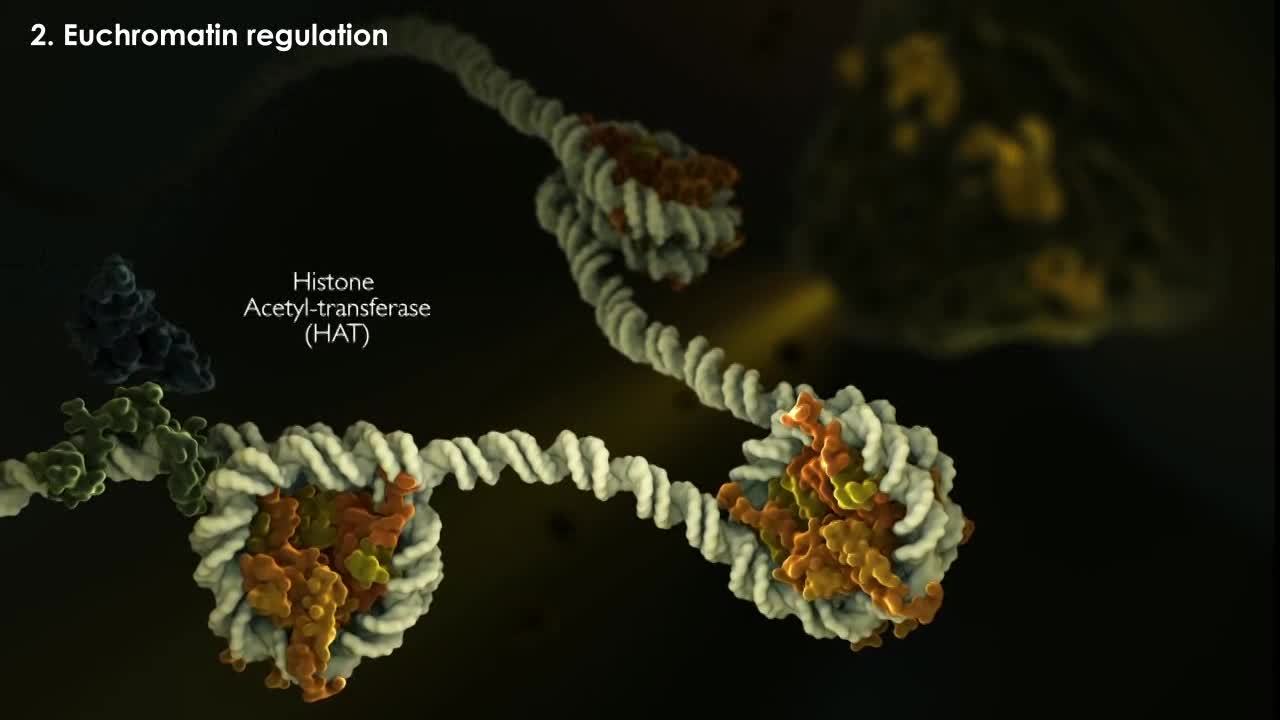
Examples of these modifications include methylation for Histone H3, acetylation for Histone H4 and phosphorylation for Histone H2B. New chromatin is often characterized by a more open and accessible state of the DNA, one in which transcription factors have access to their cognate binding sites and can therefor recruit enzymes like histone acetyltransferases that acetylate histone tails and activate genes by recruiting components of the basal transcriptional machinery including RNA polymerase.
Heterochromatin in contrast, is thought to be characterized by my repressive, tight bundling on nucleosomes which impedes transcription factors from gaining access to regulatory sites on the DNA. Methylation of cytosine bases in regions called CPG Islands is a hallmark of transcriptionally repressed heterochromatin. These methylated cytosines, in turn, recruit proteins like MECP2, methyl CPG-binding protein 2 and HP1, heterochromatic Protein 1.
These proteins are thought to maintain a repressive state of chromatin by inducing histone deacetylation by HDACs as well has histone tail methylation by histone methyltransferases enzymes.
Related Videos
-

What Is Epigenetics? -

How Complex is Epigenetics? -

How Can Epigenetics Help Us Treat Diseases?

