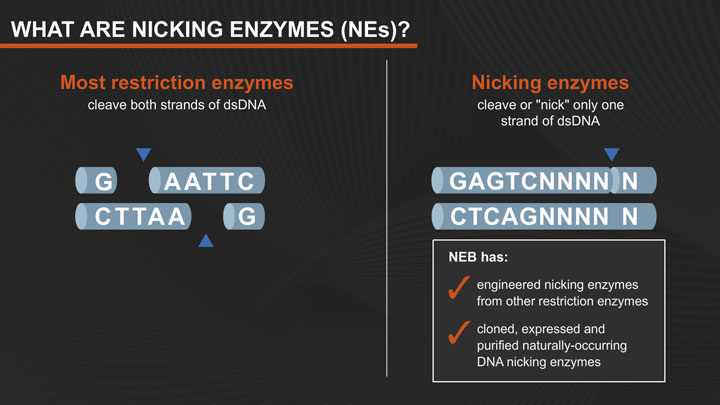
As a rule, when restriction endonucleases bind to their recognition sequences in DNA, they hydrolyze both strands of the duplex at the same time. Two independent hydrolytic reactions proceed in parallel, most often driven by the presence of two catalytic sites within each enzyme, one for hydrolyzing each strand. NEB has engineered altered restriction enzymes (nicking endonucleases) that hydrolyze only one strand of the duplex, to produce DNA molecules that are “nicked”, rather than cleaved. These conventional nicks (3´-hydroxyl, 5´-phosphate) can serve as initiation points for a variety of further enzymatic reactions such as replacement DNA synthesis, strand-displacement amplification (1), exonucleolytic degradation or the creation of small gaps (2).
There are also 3 naturally occurring nicking endonucleases: Nt.BstNBI, Nb.BtsI, and Nb.BsrDI. They are the large subunits of heterodimeric restriction enzymes. As such, the catalytic site present in the small subunit that catalyzes cleavage of the other strand is entirely missing. These 3 enzymes should therefore display absolutely no double-strand cleavage activity.
Nicking endonucleases are as simple to use as restriction endonucleases. Since the nicks generated by 6- or 7-base nicking endonucleases do not fragment DNA, their activities are monitored by conversion of supercoiled plasmids to open circles. Alternatively, substrates with nicking sites close enough on opposite strands to create a double-stranded cut can be used instead.
The uses of nicking endonuclease are still being explored. NEases can generate nicked or gapped duplex DNA for DNA mismatch repair studies and for diagnostic applications. The long overhangs that nicking enzymes make can be used in DNA fragment assembly. Nt.BbvCI has been used to generated long and non-complementary overhangs when used with XbaI in the USERTM cloning protocol from NEB. Nicking endonucleases are also useful for isothermal DNA amplification, which relies on the production of site-specific nicks. For example, isothermal DNA amplification using Nt.BstNBI in concert with Vent (exo-) DNA Polymerase (NEB #M0257) (EXPAR) has been reported for detection of a specific DNA sequence in a sample (3). Another isothermal DNA amplification technique (Nicking Endonuclease Mediated DNA Amplification (NEMDA)) (4) has been described using the 3-base cutter Nt.CviPII and Bst DNA Polymerase I. Frequent cutting NEases can generate short partial duplex DNA fragments from genomic DNA. These fragments can be used for cloning or used as probes for hybridization based applications. Nicking enzymes have also been used for genome mapping.
WarmStart Nt.BstNBI is inactive below 40°C. The WarmStart feature enables high-throughput applications, room temperature setup, and can increase the consistency and specificity of amplification reactions that utilize nicking enzymes, such as Strand Displacement Amplification (SDA) and Nicking Enzyme Amplification Reaction (NEAR).
NEB continues to engineer more nicking enzymes, particularly in response to specific customer needs and applications. Please inquire at info@neb.com.
| Enzyme |
NEB Cat# |
Description |
Reference |
| Nt.BspQI |
R0644 |
Derivative of the restriction enzyme BspQI that cleaves one strand of DNA on a double-stranded DNA substrate. |
19747545 |
| Nt.CviPII |
R0626 |
Naturally occurring nicking endonuclease cloned from chlorella virus NYs-1. |
|
| Nt.BstNBI |
R0607 |
Naturally occurring thermostable nicking endonuclease cloned from Bacillus Stereothermophilus. |
11410656
11154070 |
| WarmStart® Nt.BstNBI |
R0725 |
Naturally occurring thermostable nicking endonuclease cloned from Bacillus Stereothermophilus; formulated with a reversibly-bound aptamer, which inhibits its nicking activity at temperatures below 40°C. |
11410656
11154070
Rohloff, J.C., et al. (2014) Mol Ther Nucleic Acids. 3, e201.
Gelinas, A.D., et al. (2016) Curr Opin Struct Biol. 36, 122–132
|
| Nb.BsrDI |
R0648 |
Naturally occurring large subunit of thermostable heterodimeric enzyme |
Zhu, Z. and Xu, S.Y. unpublished results. |
| Nb.BtsI |
R0707 |
Naturally occurring large subunit of thermostable heterodimeric enzyme |
Zhu, Z. and Xu, S.Y. unpublished results. |
| Nt.AlwI |
R0627 |
Derivative of the restriction enzyme AlwI (NEB #R0513). Has been engineered to behave in the same way (6). Both nick just outside their recognition sequences. |
11687651 |
| Nb.BbvCI |
R0631 |
Derivative of the heterodimeric restriction enzyme BbvCI, engineered to possess only one functioning catalytic site |
15826660 |
| Nt.BbvCI |
R0632 |
Derivative of the heterodimeric restriction enzyme BbvCI, engineered to possess only one functioning catalytic site |
15826660 |
| Nb.BsmI |
R0706 |
Bottom-strand specific variant of BsmI (NEB #R0134) discovered from a library of random mutants |
15019778 |
| Nb.BssSI |
R0681 |
Bottom-strand specific variant of wild-type BssSI, for which we now have a more robust enzyme with lower star activity, BssSI-v2 (NEB #R0680) |
|
| Nt.BsmAI |
R0121 |
Top-strand specific variant of BsmAI (NEB #R0529) |
15019778
15247348
|

(1) Walker, G.T. et al. (1992) Proc. Natl. Acad. Sci. USA, 89, 392–396. PMID: 1309614
(2) Wang, H. and Hays, J.B. (2000) Mol. Biotechnol., 15, 97–104. PMID: 10949822
(3) Van Ness, J et al. (2003) Proc. Natl. Acad. Sci. USA, 100, 4504-4509. PMID: 12679520
(4) Chan, S l. (2004) Nucleic Acids Res., 32, 6187-6188. PMID: 15570069