DNase I (RNase-free)
GMP-grade reagent also available. Learn more.
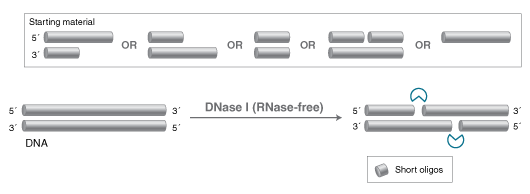
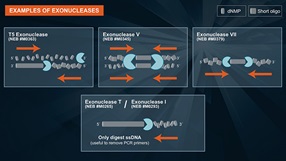
- DNA specific Endonuclease
- Degrades double-stranded and single-stranded DNA
- Products are short oligos with 5'-phosphate and 3'-OH
- Need help finding the right exonuclease for your experiments? Try Exo Selector
DNase I (RNase-free) is ideal for:
- Removal of contaminating genomic DNA from RNA samples
- Degradation of DNA templates in transcription reactions
- Salt-tolerant DNase I-XT (NEB #M0570) is now available for removal of DNA from RNA preparations

-
Product Information
DNase I, (RNase-free) is an endonuclease that nonspecifically cleaves DNA to release di-, tri- and oligonucleotide products with 5´-phosphorylated and 3´-hydroxylated ends (1,2). DNase I acts on single- and double-stranded DNA, chromatin and RNA:DNA hybrids. Highlights
Isolated from a recombinant source
Supplied with 10X Reaction BufferProduct Source
An E. Coli strain that carries an MBP fusion clone of Bovine Pancreatic DNase I.- This product is related to the following categories:
- Exonucleases and Non-specific Endonucleases Products,
- RNA Synthesis In vitro Transcription (IVT)
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.