-
Product Information
Product Source
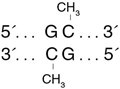
The GpC Methyltransferase, M.CviPI, is isolated from a strain of E. coli which contains the methyltransferase gene from Chlorella virus. This construct is fused to the maltose binding protein (MBP).- This product is related to the following categories:
- Methyltransferases for Epigenetics Products,
- Methyltransferases Products
-
Protocols, Manuals & Usage
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.