Uracil-DNA Glycosylase (UDG)
UDG has been reformulated with Recombinant Albumin (rAlbumin) beginning with Lot #10225520. Learn more.
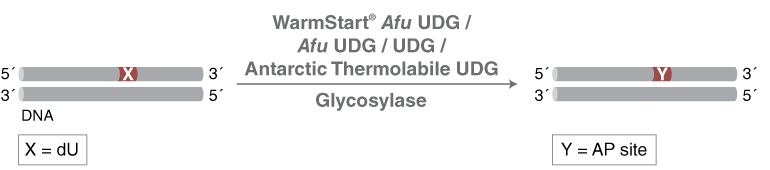
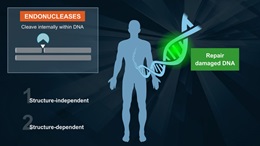
- Uracil DNA glycosylase
- Monofunctional DNA glycosylase that catalyzes the hydrolysis of the N-glycosidic bond from deoxyuridine to release uracil
- Active on ss and dsDNA
- Heat inactivation not possible
Featured Video
-
Product Information
E. coli Uracil-DNA Glycosylase (UDG) catalyses the release of free uracil from uracil-containing DNA. UDG efficiently hydrolyzes uracil from single-stranded or double-stranded DNA, but not from oligomers (6 or fewer bases).
Product Source
An E. coli strain that carries the UDG gene from E. coli.- This product is related to the following categories:
- DNA Repair Enzymes and Structure-specific Endonucleases Products
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.