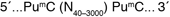McrBC
- Catalog # M0272 was discontinued on September 15, 2023
McrBC is an endonuclease which cleaves DNA containing methylcytosine* on one or both strands. Sites on the DNA recognized by McrBC consist of two half-sites of the form (G/A)mC. These half-sites can be separated by up to 3 kb, but the optimal separation is 55-103 base pairs.
*5-methylcytosine or 5-hydroxymethylcytosine
- McrBC requires GTP for DNA cleavage
- Supplied with NEBuffer 2, Recombinant Albumin and 100x GTP

-
Product Information
Recognition Site:
McrBC is an endonuclease which cleaves DNA containing methylcytosine* on one or both strands. McrBC will not act upon unmethylated DNA (1). Sites on the DNA recognized by McrBC consist of two half-sites of the form (G/A)mC. These half-sites can be separated by up to 3 kb, but the optimal separation is 55-103 base pairs (2,3). McrBC requires GTP for cleavage, but in the presence of a non-hydrolyzable analog of GTP, the enzyme will bind to methylated DNA specifically, without cleavage (4).
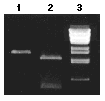
*5-methylcytosine or 5-hydroxymethylcytosine (5).Linearized plasmid (methylated or unmethylated), containing one McrBC site, incubated with McrBC. Lane 1, unmethylated DNA; Lane 2, methylated DNA; Lane 3, Marker: Lambda DNA BstEII digest.
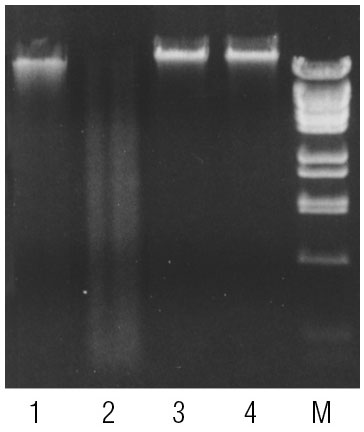
Human and Drosophila genomic DNA incubated with McrBC. 60-90% of CpGs in vertebrate DNA are estimated to be methylated (6) while methylated CpGs are extremely rare in Drosophila DNA (7). Lane 1, Human DNA; Lane 2, Human DNA incubated with McrBC; Lane 3, Drosophila DNA; Lane 4, Drosophila DNA incubated with McrBC; Lane 4, Marker: Lambda DNA BstEII digest. Product Source
The two component proteins are purified separately from E. coli K-12 strains containing plasmids encoding McrB and McrC (1).- This product is related to the following categories:
- Discontinued (<3 years)
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.