Navigating mRNA IVT
Posted on Monday, March 25, 2024
By
Topic: Tips for the lab, What is Trending in Science
Harnessing the full potential of mRNA technology hinges upon the molecular strategies that you select for in vitro transcribed synthesis. NEB has decades of experience developing best-in-class reagents for RNA research and has partnered with leading biopharmaceutical companies for global-scale manufacturing of mRNA vaccines. This overview of in vitro transcription (IVT) describes opportunities at each step to steer you toward effective mRNA synthesis. We invite you to download our new E-book, From Template to Transcript, Overcoming Challenges in mRNA Manufacturing, for more in-depth insights.
In vitro transcribed mRNA has proven its value for both basic research and the rapid development of lifesaving therapeutics, thanks to its relatively simple manufacturing process. IVT mRNA synthesis is a flexible, cell-free, and rapidly scalable method. The main steps proceed from template preparation through to RNA synthesis, modification, and finally, purification. Each step offers options to improve workflow efficiency and the ultimate quality of your mRNA molecules. Anticipating a path for scaling can also be important. Early-stage therapeutic studies often require vast amounts of RNA. IVT mRNA production can be scaled up or scaled out with GMP-grade mRNA manufacturing materials. mRNA IVT technology has changed the landscape of drug research, development, and production. Your discoveries can be put into effect faster.
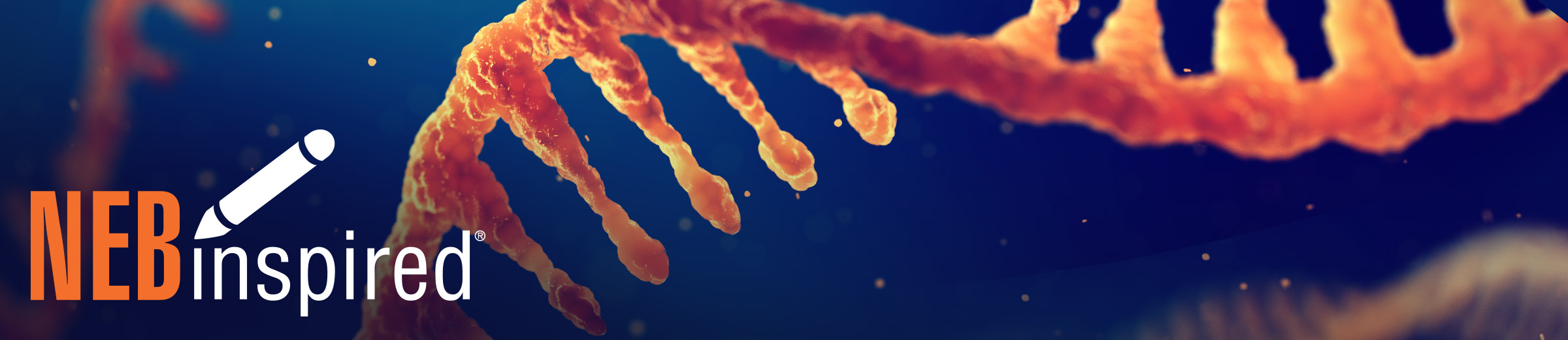
Choosing Tactics for IVT Templates
The ultimate quality of your mRNA molecules and in vitro synthesis efficiency begins with your template. The foundation of any IVT — the dsDNA template — contains a promoter upstream of your gene target and 5′ untranslated region (UTR) to facilitate ribosome recruitment. Template sequence engineering can improve mRNA translational capacity in vivo, as well as in vitro transcription and modification efficiencies. You can choose to generate your template with a plasmid-based strategy or an enzymatic amplification-based strategy. Iteration on template design can be effectively accomplished using our cloning tools, including the NEBuilder® HiFi DNA Assembly and NEBridge® Golden Gate Assembly, in conjunction with our online cloning resources. Regardless of your approach for template generation, a precisely encoded poly(A) sequence in your DNA template will limit untemplated extensions and potential double-stranded RNA byproducts.
Here are a few of our recommendations to generate error-free, fully linearized, DNA template for IVT at high yields:
Plasmid linearization
You can simplify the rapid construction of complex plasmids for IVT templates by using the right DNA enzymes and digital design tools for DNA assembly. Plasmid linearization can be optimized with your selection of restriction enzyme and digest conditions - as discussed in our previous blog post on choosing a restriction enzyme for mRNA vaccine production. You can plan to produce a scarless template with 5´-overhanging ends or blunt ends that allow downstream RNA synthesis with no additional 3´-nucleotide sequence from the restriction site – all while maintaining the poly(A) tail.
Polymerase choice
Minimizing errors when generating synthetic DNA templates with PCR amplification relies on the choice of DNA polymerase and reaction conditions. The Q5® High-Fidelity DNA Polymerase (NEB #M0491) is composed of a novel polymerase that is fused to the processivity-enhancing Sso7d DNA binding domain and has a ~280X lower error rate than Taq for a broad range of amplicons. Q5 is also available in hot start and master mix formats. Alternatively, you can produce templates through isothermal amplification approaches such as rolling circle amplification-based methods. Here, errors can be minimized using phi29 DNA Polymerase (NEB #M0269) for its exceptional strand displacement and processive synthesis properties and inherent 3'→5' proofreading exonuclease activity. Whether you use PCR or isothermal amplification, choosing the right polymerase can increase accuracy, reduce workflow times, and increase template yield.
Template purification
Purification of the template is critical for successful IVT. After plasmid linearization or amplification, Monarch® PCR and DNA Cleanup Kits can be used to remove enzymes and reaction reagents. Monarch's unique column design enables elution in as little as 6μl and prevents buffer/salt carryover. Clean RNA can be obtained in minutes with quick and simple protocols.
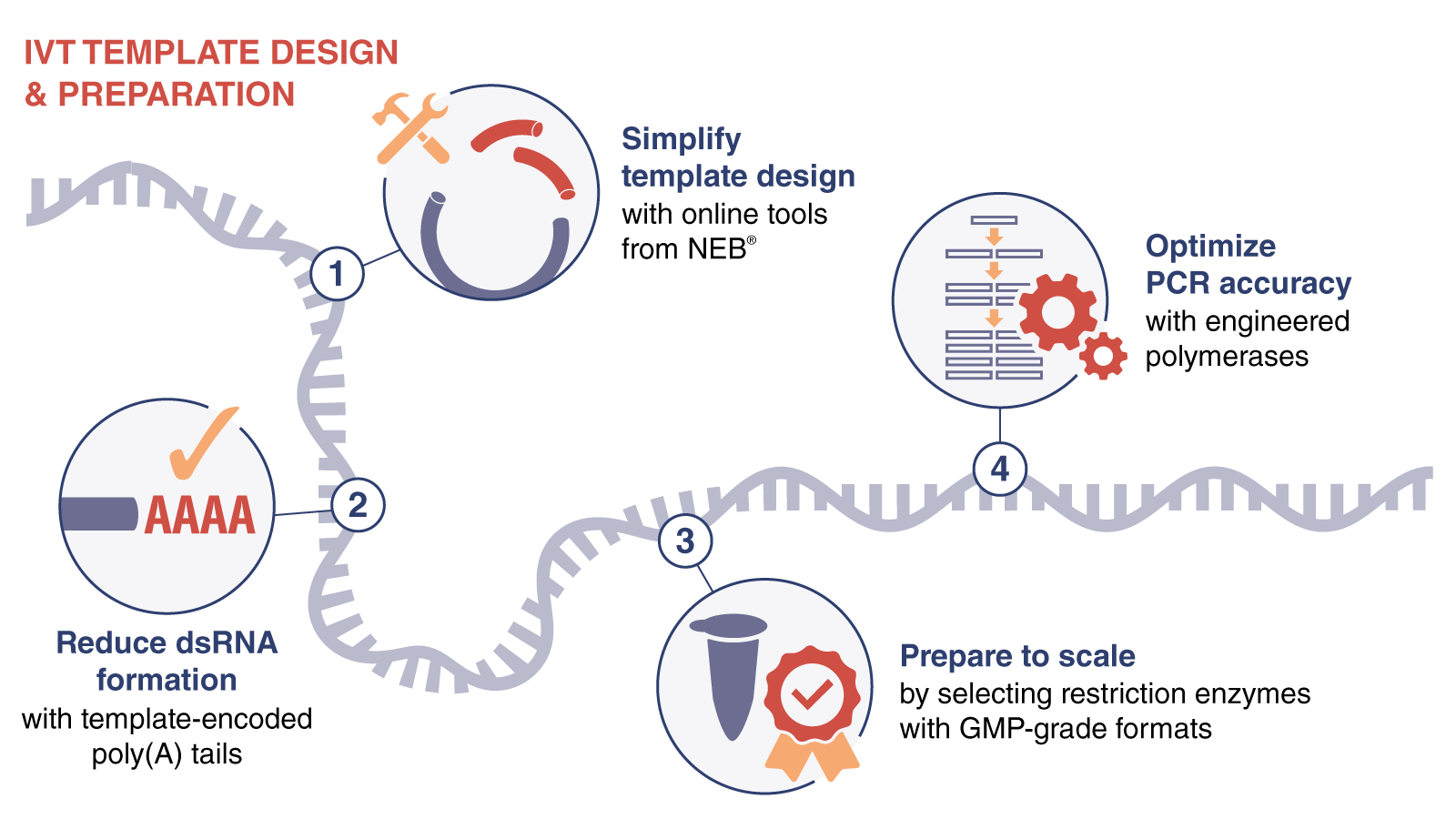
Setting the Course for Efficient RNA Synthesis, Capping, and Tailing
Production of functional mRNA requires the incorporation of unique elements at both termini. At the 5’ end, the 7-methylguanylate cap structure can be generated post-transcriptionally using capping enzymes or co-transcriptionally using cap analogs. The 3’ end is extended via polyadenylation, or “tailing”, and can also be produced enzymatically using poly(A) polymerase or encoded in the template. The nucleotide sequence of mRNA has a great influence on effective IVT and 5′ capping. Additionally, the scale of your IVT and whether you plan to incorporate modified nucleotides at this stage are also critical determinants.
We recommend enzyme-based post-transcriptional capping for large-scale mRNA synthesis. Following transcription, the 5' end is first converted to Cap-0 with Faustovirus or Vaccinia Capping Enzyme (NEB #M2080), then is converted to Cap-1 by 2’-O-methylation of the first RNA ribose using mRNA cap 2´-O-methyltransferase (MTase) (NEB #M0366). Increasing the IVT reaction temperature with standard T7 RNA Polymerase or using the thermostable RNA polymerase Hi-T7 RNA Polymerase (NEB #M0658) can reduce 3′ extension of run-off products. Faustovirus Capping Enzyme, known as FCE, (NEB #M2081) requires less enzyme to cap most transcripts and has a wide temperature range making it a good choice for long RNA or self-amplifying RNAs, or other difficult-to-cap substrates.
As you start to engage in exploratory research, our robust IVT kits provide high yields with minimal optimization across templates. NEB HiScribe® Kits for IVT are compatible with post-transcriptional or co-transcriptional capping approaches. Poly(A) can be added through an encoded poly(A) tail or enzymatically with E.coli Poly(A) Polymerase (NEB #M0276). These are great starting points for a wide range of template lengths with streamlined workflows and minimal optimization required. In particular, the HiScribe T7 mRNA Kit with CleanCap® Reagent AG (NEB #E2080) is ideal for generating a multitude of capped transcripts for screening. Co-transcriptional capping with trinucleotide cap analogs has made waves by dramatically improving the capping efficiency and yields relative to first-generation cap analogs such as Anti-Reverse Cap Analog (ARCA). You can generate greater than 95% Cap-1 mRNA in a single reaction with CleanCap Reagent AG.
NEB offers both high-yield HiScribe kits and individual enzymes at research grade or GMP grade* when you’re scaling up or scaling out. These reagents are functionally equivalent, allowing optimization at low volumes with research-grade enzymes before scaling up your process for commercial and clinical production with GMP-grade materials.
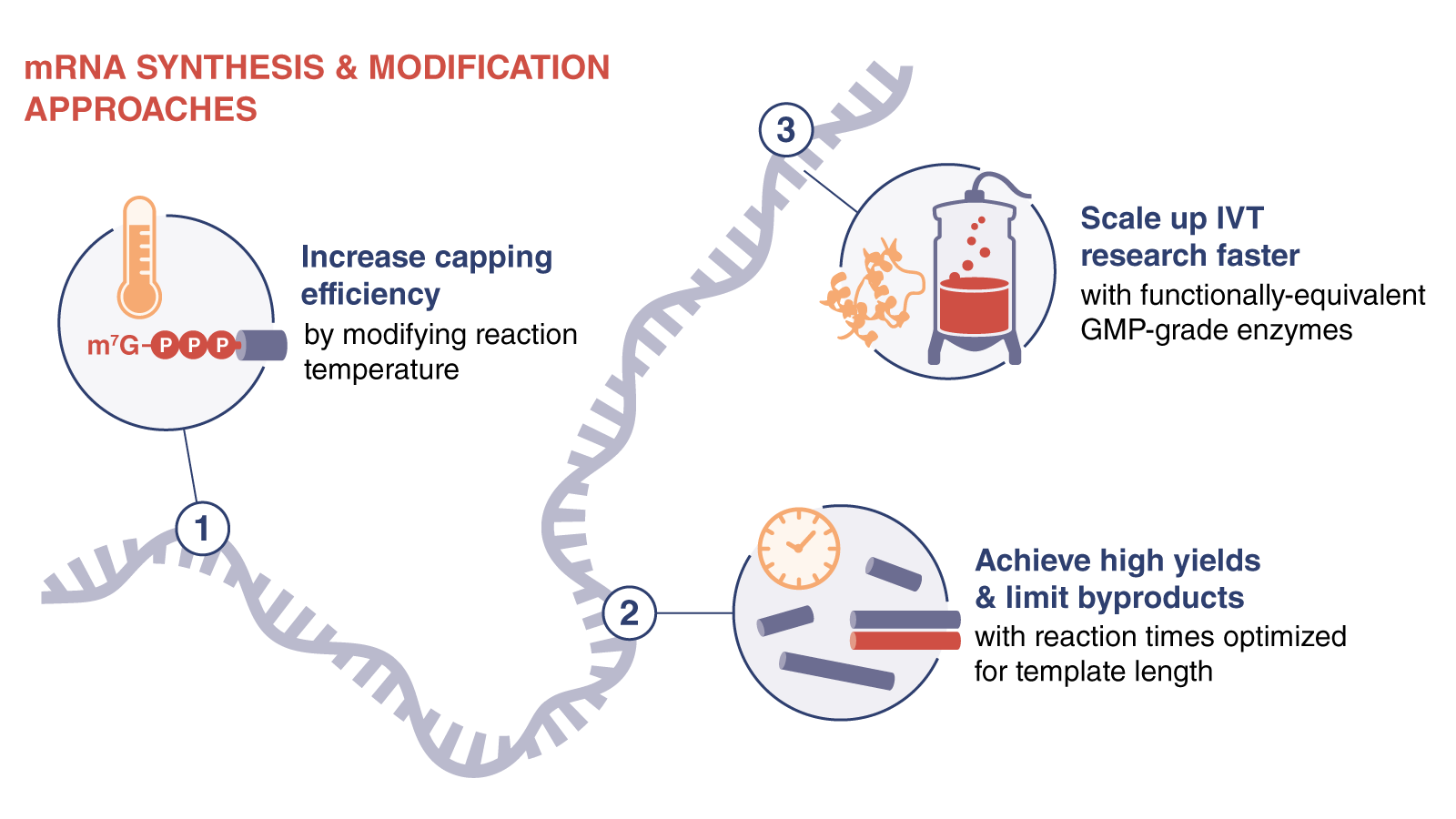
Purifying your mRNA
After synthesis and modification, your mRNA must be cleaned up to remove impurities. At this point, you have options to simplify downstream processing. The simplest way to remove template from your reaction is with a DNase I digest. You can skip adjusting the buffer if you use DNase I-XT (NEB #M0570), which NEB scientists engineered to remain active in solutions containing up to 300 mM salt. You can remove contaminants to ready your mRNA for RT-PCR, RNA library prep for NGS, RNA labeling, RNAi, microinjection and or transfection by using our Monarch RNA Cleanup Kits in a quick spin-column format. These are ideal for cleanup and concentration of high quality, high purity mRNA after enzymatic reactions.
Tap our IVT expertise for high quality mRNA
We hope this overview of the steps to effectively synthesize RNA is helpful. If you are seeking a partner with technical expertise, pressure-tested production capacity and regulatory experience in mRNA-based therapeutics, please contact our OEM & Customized solutions team.
Download eBook
NEB will not rent, sell or otherwise transfer your data to a third party for monetary consideration. See our Privacy Policy for details. View our Community Guidelines.
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. See www.neb.com/trademarks. The use of these products may require you to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
Don’t miss out on our latest NEBinspired blog releases!
- Sign up to receive our e-newsletter
- Download your favorite feed reader and subscribe to our RSS feed
Be a part of NEBinspired! Submit your idea to have it featured in our blog.



