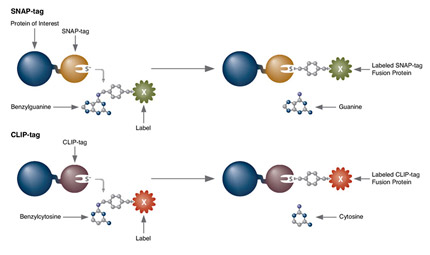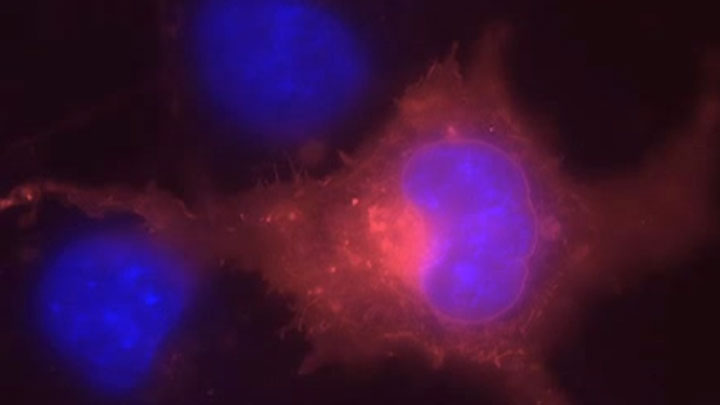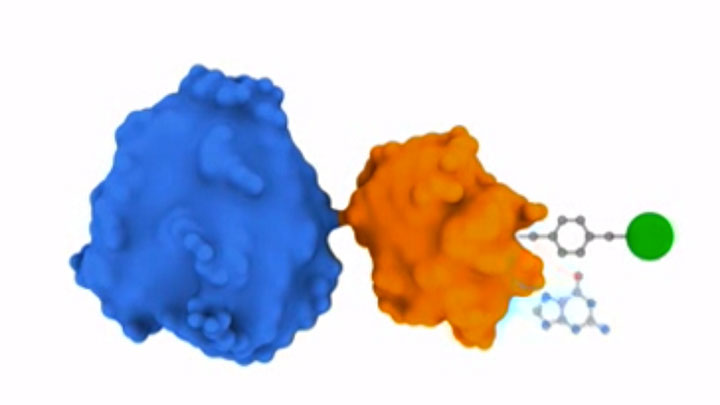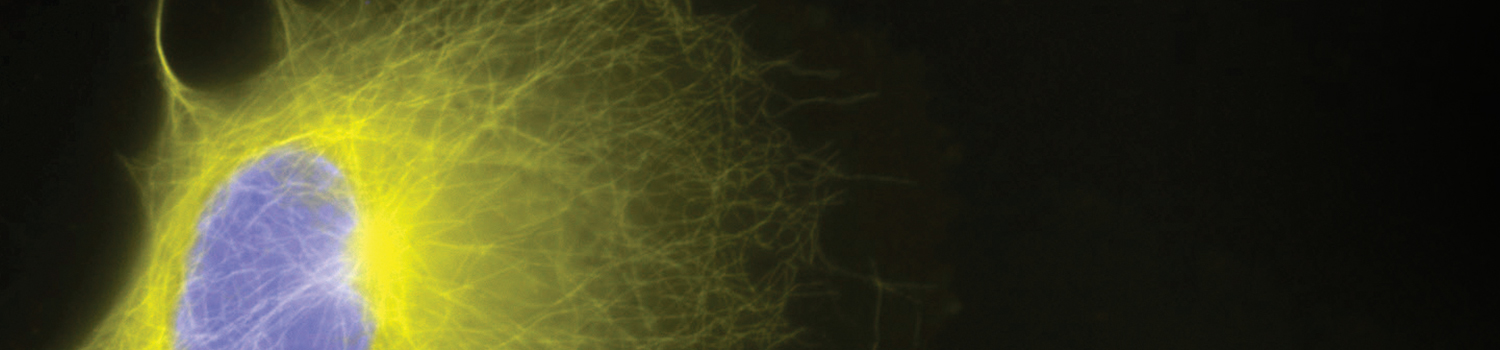
SNAP Surface
Choose Type:
- Cellular Labeling (E9100)
- Cellular Labeling (S9103)
- Cellular Labeling (S9105)
- Cellular Labeling (S9107)
- Cellular Labeling (S9109)
- Cellular Labeling (S9110)
- Cellular Labeling (S9112)
- Cellular Labeling (S9124)
- Cellular Labeling (S9129)
- Cellular Labeling (S9132)
- Cellular Labeling (S9134)
- Cellular Labeling (S9136)
- Cellular Labeling (S9101)
- Labeling of Proteins in vitro (S9110)
- Labeling of Proteins in vitro (S9103)
- Labeling of Proteins in vitro (S9105)
- Labeling of Proteins in vitro (S9107)
- Labeling of Proteins in vitro (S9109)
- Labeling of Proteins in vitro (S9143)
- Labeling of Proteins in vitro (S9101)
- Labeling Proteins in vitro (S9112)
- Labeling Proteins in vitro (S9124)
- Labeling Proteins in vitro (S9129)
- Labeling Proteins in vitro (S9132)
- Labeling Proteins in vitro (S9134)
- Labeling Proteins in vitro (S9136)
- Use of SNAP-Cell Block with SNAP-Cell Substrates (E9100)
- View the video "Fluorescent Labeling of COS-7 Expressing SNAP-tag Fusion Proteins for Live Cell Imaging" in the Journal of Visualized Experiments (JoVE)
- Cellular Labeling (E9120)
- Labeling Proteins in vitro (E9120)
- Cloning of SNAP-tag Fusions in pSNAPf (N9183)
- Cloning of SNAP-tag Fusions in pSNAP-tag(T7)-2 (N9181)
- Expression of SNAP-tag Fusions (N9181)
- Expression of SNAPf Fusions (N9183)
- Labeling of Proteins in vitro (E9100)
- Use with SNAP-Surface substrates (S9143)
- Cellular Labeling (S9159)
- Labeling of Proteins in vitro (S9159)
-
SNAP-tag® Technologies: Tools to Study Protein Function
Read about the NEB’s set of protein tools for the specific labeling (SNAP-, CLIP-, ACP- and MCP-tags) of fusion proteins.
- Cellular Imaging & Analysis Brochure
- Comparison of SNAP-tag®/CLIP-tag™ Technologies to GFP
- SNAP-tag® and CLIP-tag™ Substrate Selection Chart
- Labeling with SNAP-tag® Technology Troubleshooting Guide
Feature Articles
Brochures
Selection Tools
Troubleshooting Guides
- Maffei, M., Morelli, C., Graham, E., Patriarca, S., Donzelli, L., Doleschall, B., de Castro, Reis, F., Nocchi, L., Chadick, C.H., Reymond, L., Correa, I.R., Jr., Johnsson, K., Hackett, J.A., Heppenstall, P.A (2019) A ligand based system for receptor specific delivery of proteins Sci Rep; 9(1), 19214.. PubMedID: 31844114, DOI: 10.1038/s41598-019-55797-1
- Meron Mengistu, Krishanu Ray, George K Lewis, Anthony L DeVico (2015) Antigenic properties of the human immunodeficiency virus envelope glycoprotein gp120 on virions bound to target cells PLoS Pathog; 11(3), e1004772. PubMedID: 25807494, DOI: 10.1371/journal.ppat.1004772.
- Gabriele Fuchs, Alexey N Petrov, Caleb D Marceau, Lauren M Popov, Jin Chen, Sen E O'Leary, Richard Wang, Jan E Carette, Peter Sarnow, Joseph D Puglisi (2015) Kinetic pathway of 40S ribosomal subunit recruitment to hepatitis C virus internal ribosome entry site Proc Natl Acad Sci U S A; 112(2), 319-325. PubMedID: 25516984, DOI: 10.1073/pnas.1421328111.
- Elena Shvets, Vassilis Bitsikas, Gillian Howard, Carsten Gram Hansen, Benjamin J Nichols (2015) Dynamic caveolae exclude bulk membrane proteins and are required for sorting of excess glycosphingolipids Nat Commun; 6, 6867. PubMedID: 25897946, DOI: 10.1038/ncomms7867.
- Margaret L Rodgers, Joshua Paulson, Aaron A Hoskins (2015) Rapid isolation and single-molecule analysis of ribonucleoproteins from cell lysate by SNAP-SiMPull RNA; 21(5), 1031-1041. PubMedID: 25805862, DOI: 10.1261/rna.047845.114
- Juri Nio Bach, Marc Bramkamp (2015) Dissecting the molecular properties of prokaryotic flotillins PLoS One; 10(1), e0116750. PubMedID: 25635948, DOI: 10.1371/journal.pone.0116750.
- Clone and express once, then use with a variety of substrates
- Non-toxic to living cells
- Wide selection of fluorescent substrates
- Highly specific covalent labeling
- Simultaneous dual labeling
- Simultaneous dual protein labeling on the surface of live cells
- Protein localization and translocation
- Pulse-chase experiments
- Receptor internalization studies
- Selective cell surface labeling
- Protein pull-down assays
- Protein detection in SDS-PAGE
- Flow cytometry
- High throughput binding assays in microtiter plates
- Biosensor interaction experiments
- FRET-based binding assays
- Single molecule labeling
- Super-resolution microscopy
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.