Endonuclease V
The large size of this product has been discontinued.

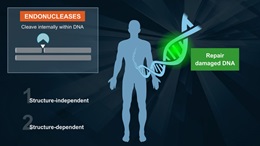
- DNA endonuclease
- Catalyzes the cleavage of the second DNA phosphodiester backbone 3' to deoxyinosine, leaving a 3'-OH and 5' - phosphate
- Endonuclease V activity, to a lesser extent, is also reported on AP site, urea, mismatches, hairpins, loops, and pseudo-Y structures
Featured Video
-
Product Information
Endonuclease V is a repair enzyme found in E. coli that recognizes deoxyinosine, a deamination product of deoxyadenosine in DNA. Endonuclease V, often called deoxyinosine 3´ endonuclease, recognizes DNA containing deoxyinosines (paired or not) on double-stranded DNA, single-stranded DNA with deoxyinosines and to a lesser degree, DNA containing abasic sites (ap) or urea, base mismatches, insertion/deletion mismatches, hairpin or unpaired loops, flaps and pseudo-Y structures. It is believed that Endonuclease V needs another protein to repair the DNA, as it does not remove the deoxyinosine or the damaged bases (1,2,3).
Endonuclease V cleaves the second phosphodiester bonds 3´ to the mismatch of deoxyinosine (2), leaving a nick with 3´-hydroxyl and 5´-phosphate (4).
Product Source
An E. coli strain containing a gene fusion of the Endo V gene and the gene coding for the maltose binding protein (MBP). The fusion protein is purified to near homogeneity and is active as a fusion. The protein contains 223 amino acids and has a molecular weight of 24.9 kDa (5).- This product is related to the following categories:
- DNA Repair Enzymes and Structure-specific Endonucleases Products
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.