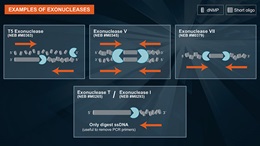
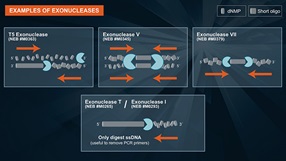
Exonuclease T

- Also known as RNase T
- Single-stranded (ssDNA or RNA) specific exonuclease
- Catalyzes the removal of nucleotides from linear single-stranded DNA or RNA in the 3' to 5' direction
- Need help finding the right exonuclease for your experiments? Try Exo Selector
Exonuclease T can be used for:
- Removal of 3' overhangs of double-stranded DNA to generate blunt-ends*
- Removal of single-stranded primers in PCR reactions prior to DNA sequencing or SNP analysis
- Removal of single-stranded primers for nested PCR reactions
- Removal of single-stranded DNA, leaving behind double-stranded DNA in the sample
- Not sure which RNase to choose? Learn more about the different RNases offered by NEB.
*sequence-dependent
Featured Video
-
Product Information
Exonuclease T (Exo T), also known as RNase T, is a single-stranded RNA (1,2) or DNA (3,4) specific nuclease that requires a free 3´ terminus and removes nucleotides in the 3´→ 5´ direction. Exonuclease T can be used to generate blunt ends from RNA (5) or DNA molecules that have 3´ extensions (4). Product Source
Exonuclease T is overexpressed and purified as a C-terminal fusion to maltose-binding protein (MBP). MBP is removed from Exonuclease T by Factor Xa cleavage and Exonuclease T is then purified away from Factor Xa and MBP. Exonuclease T cleaved from MBP has an additional amino acid on the N-terminus and a Phe instead of a Met (i.e., Glu-Phe-Exo T instead of Met-Exo T).- This product is related to the following categories:
- RNases,
- Exonucleases and Non-specific Endonucleases Products
-
Tools & Resources
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.