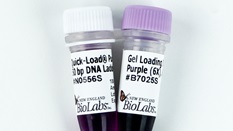
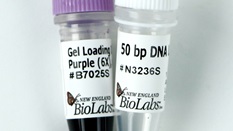
Low Molecular Weight DNA Ladder
This product is available in our Quick-Load Purple format - enjoy the convenience of a ready-to-load format.
- Size range: 25 bp to 766 bp
- Value pricing
- Supplied with free vial of Gel Loading Dye, Purple (6X), no SDS (NEB #B7025).
- Easy to identify reference bands
- Small size suitable for 100 gel lanes; large size suitable for 500 gel lanes

-
Product Information
A proprietary plasmid is digested to completion with appropriate restriction enzymes to yield 11 bands suitable for use as molecular weight standards for both agarose and polyacrylamide gel electrophoresis. This digested DNA includes fragments ranging from 25-766 base pairs. The 200 base pair band has increased intensity to serve as a reference point. Comes supplied with 1 vial of Gel Loading Dye, Purple (6X), no SDS.

- This product is related to the following categories:
- DNA Markers & Ladders Products
- This product can be used in the following applications:
- DNA Analysis
-
Protocols, Manuals & Usage
-
Tools & Resources
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.