RecA
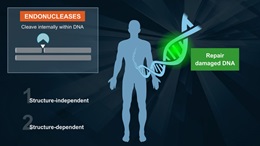
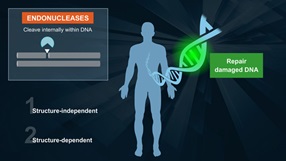
RecA is necessary for genetic recombination reactions involving DNA repair and UV-induced mutagenesis
- Visualization of DNA structures with electron microscopy
- D-loop mutagenesis
- Screening libraries using RecA-coated probes
- Cleavage of DNA at a single predetermined site
- RecA mediated affinity capture for full length cDNA cloning
Featured Video
-
Product Information
E. coli RecA is necessary for genetic recombination, reactions involving DNA repair and UV-induced mutagenesis. RecA promotes the autodigestion of the lexA repressor, umuD protein and lambda repressor. Cleavage of LexA derepresses more than 20 genes (1). In vitro studies indicate that in the presence of ATP, RecA promotes the strand exchange of single-strand DNA fragments with homologous duplex DNA. The reaction has three distinct steps: (i) RecA polymerizes on the single-strand DNA, (ii) the nucleoprotein filament binds the duplex DNA and searches for a homologous region, (iii) the strands are exchanged (2).
- This product is related to the following categories:
- ssDNA Binding Proteins Products,
- DNA Repair Enzymes and Structure-specific Endonucleases Products
-
Protocols, Manuals & Usage
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Featured Videos
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.