The Structure, Function and Importance of Carbohydrates
Glycobiology is the study of the structure, function and biology of carbohydrates, also called glycans, which are widely distributed in nature. It is a small but rapidly growing field, with relevance to biomedicine, biotechnology and basic research. Proteomics, the systematic study of proteins in biological systems, has expanded the knowledge of protein expression, modification, interaction and function. However, in eukaryotic most proteins are post-translationally modified.
Key Functions of Carbohydrates
(1). A common post-translational modification essential for cell viability is the attachment of glycans, as shown in Figure 1. Glycosylation defines the adhesive properties of glycoconjugates and it is largely through glycan–protein interactions that cell– cell and cell–pathogen contacts occur, a fact that highlights the importance of glycobiology. Considering the central role that glycans play in molecular encounters, glycoprotein and carbohydrate-based drugs and therapeutics represent a greater than $20 billion market
(2). Glycomics, the systematic study of all glycan structures in a biological system, relies on effective enzymatic and analytical techniques for correlation of glycan structure with function.
Classification of Glycans
Several classes of glycans exist, including N-linked glycans, O-linked glycans, glycolipids, O-GlcNAc, and glycosaminoglycans. N-linked glycosylation occurs when glycans are attached to asparagine residues on the protein. O-linked glycans are most commonly attached to serine or threonine residues through the N-Acetylgalactosamine residue. Removal of oligosaccharides from glycoproteins, termed deglycosylation, is often used in order to simplify analysis of the peptide and/ or glycan portion of a glycoprotein. Detailed knowledge of the glycan structures helps to correlate them to their respective function. To do this, tools are required for highly sensitive analysis of glycan chains.
Both chemical and enzymatic methods exist for removing oligosaccharides from glycoproteins. Chemical methods such as β-elimination with mild alkali (3) or mild hydrazinolysis (4) result in the degradation of the protein; whereas enzymatic methods are much gentler and can provide complete sugar removal with no protein degradation.
Figure 1: N- and O-Glycosylation
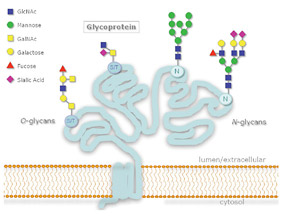
protein, while O-linked glycosylation occurs through serine or threonine.
N-Linked Glycans
For structural analysis of asparagine-linked carbohydrates, sugars are released from the protein backbone by enzymes such as PNGase F (NEB #P0704). PNGase F is the most effective enzyme for removing almost all N-linked oligosaccharides from glycoproteins. PNGase F cleaves between the innermost GlcNAc and asparagine residues of high mannose, hybrid and complex oligosaccharides from N-linked glycoproteins (5), as shown in Figure 2.
Figure 2: Specificity of PNGase F
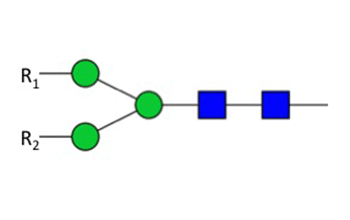
PNGase F digestion deaminates the asparagine residue to aspartic acid, leaving the oligosaccharide intact for further analysis. However, it is critical to note that oligosaccharides containing a fucose α(1-3)-linked to the glycan core often found in plant and insect glycoproteins are resistant to PNGase F, and would therefore require PNGase A treatment. Other commonly used endoglycosidases such as Endoglycosidase H (NEB #P0702) are not suitable for general deglycosylation of N-linked sugars, because it only deglycosylates glycoproteins containing primarily high mannose N-linked structures, as well as leaving one N-acetylglucosamine residue attached to the asparagine.
To achieve complete removal of N-linked glycans from a protein using PNGase F, it is recommended that the glycoprotein first be denatured by heating with SDS and DTT prior to PNGase F treatment. Denaturation of the glycoprotein will decrease the steric hindrances that can inhibit PNGase F activity. However, if denaturation of a glycoprotein is not desirable, native conditions may be used. Under native conditions PNGase F retains full activity; however more enzyme may be needed to achieve complete deglycosylation.
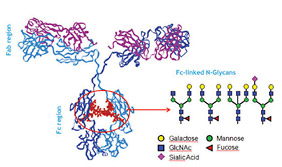
The study of N-glycosylation is related to many important pathways. Because N-glycans are added as polypeptides that are synthesized in the ER, these sugars control the correct conformation of glycoproteins. Once a correctly folded glycoprotein leaves the ER, its N-glycans are re-modeled in a protein- and cell-specific fashion. Differences in N-glycan structure result in different functions for the same polypeptide. This is well illustrated in the case of immunoglobulin G (IgG), in which the glycan composition affects complement binding, activation and other biological properties (Figure 3). Accordingly, the manufacturing of therapeutic antibodies demands assessment of a product’s microheterogeneity and its batch-to-batch consistency (6,7).
Figure 3: Typical Fc-linked glycans present in immunoglobulins
O-Linked Glycans
Removing O-linked glycan chains while rendering a protein intact for further examination is a more difficult task. Chemical methods, such as β-elimination, may result in incomplete sugar removal and degradation of the protein. On the other hand, enzymatic removal of O-linked glycans must be performed as a series of exoglycosidase digestions until only the Galβ1-3GalNAc (Core 1) and/or the GlcNAc β1-3GalNAc (Core 3) cores remains attached to the serine or threonine residue. NEB’s Enterococcus faecalis Endo-α-N-Acetylgalactosaminidase (NEB #P0733), also known as O-Glycosidase, catalyzes the removal of Core 1 and Core 3 disaccharide structures with no modification of the serine or threonine residues.
Any modification of the core structures, including sialyation, will block the action of the O-Glycosidase. Sialic acid residues are easily removed by a general α2-3,6,8 Neuraminidase (NEB #P0720). In addition, exoglycosidases such as β(1-4) Galactosidase (NEB #P0730) and β-N-Acetylglucosaminidase (NEB #P0732) can be included in deglycosylation reactions to remove other complex modifications often known to be present on the core structures.
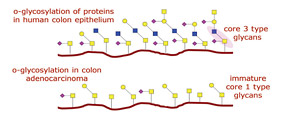
The study of O-glycosylated proteins is essential to understand the underlying mechanisms of several cellular processes. For instance, in many cancers of the mucosa (such as colon, ovary, uterus and bladder), tumor progression strongly correlates with alterations in the patterns of mucin (a surface protein) O-glycosylation (Figure 4). The enzyme responsible for Core 3 O-glycan synthesis, β3Gn-T6, is abundant in normal colon tissue while its expression is strongly downregulated in adenocarcinoma (8,9,10). As a result, mucin glycosylation switches from common Core 3 O-glycan structures to short Core 1 structures. These Core 1 structures, T and Tn, are hallmark epitopes of cell malignancy (11).
Using endoglycosidase and exoglycosidase enzymes, researchers are able to obtain highly sensitive analysis of both protein and glycan components in a wide range of healthy and diseased glycoproteins.
Figure 4: O-Glycosylation of Proteins
Learn More
Glossary
Glycobiology and Carbohydrate Related Terms
Glycobiology – the study of the structure,
function and biology of carbohydrates.
Glycomics – the systematic study of all glycan
structures in a biological system.
Carbohydrate – A generic term used interchangeably
with sugar and glycan. This term
includes monosaccharides, oligosaccharides,
and polysaccharides.
Glycan – A generic term for any sugar, in free
form or attached to another molecule, used
interchangeably with carbohydrate.
Complex glycan – A glycan containing more
than one type of monosaccharide.
β-elimination – The cleavage of a C-O or C-N
bond positioned on the β-carbon with respect
to a carbonyl group. The process is used to
cleave O-glycans from serine or threonine
residues.
Endoglycosidase – An enzyme that catalyzes
the cleavage of an internal glycosidic linkage in
an oligosaccharide or polysaccharide.
Exoglycosidase – An enzyme that cleaves
a monosaccharide from the non-reducing
end of an oligosaccharide, polysaccharide or
glycoconjugate.
Glycoconjugate – A molecule in which one
or more glycan units are covalently linked to a
non-carbohydrate entity.
Glycoforms – Different molecular forms of a
glycoprotein, resulting from variable glycan
structure and/or glycan attachment site
occupancy.
Glycoforms – Different molecular forms of a
glycoprotein, resulting from variable glycan
structure and/or glycan attachment site
occupancy.
Glycopeptide – A peptide having one or more
covalently attached glycans.
Glycoprotein – A protein with one or more
covalently attached glycans.
Glycoproteomics – The systems-level analysis
of glycoproteins, including their protein
identities, sites of glycosylation and glycan
structures.
Glycosaminoglycans – Polysaccharide side
chains of proteoglycans or free complex
polysaccharides composed of linear disaccharide
repeating units each composed of a hexosamine
and a hexose or a hexuronic acid.
Glycosylation – The enzyme-catalyzed covalent
attachment of a carbohydrate to a polypeptide,
lipid, polynucleotide or another carbohydrate,
generally catalyzed by glycosyltransferases.
Hexosamine – Hexose with an amino group in
place of the hydroxyl group at the C-2 position.
Common examples are the N-acetylated sugars,
N-acetylglucosamine and N-acetylgalactosamine.
Hexose – A six-carbon monosaccharide typically
with an aldehyde at the C-1 position and
hydroxyl groups at all other positions. Common
examples are mannose, glucose and galactose.
Hydrazinolysis – A chemical method that
uses hydrazine to cleave amide bonds (e.g.,
the glycosylamine linkage between a glycan
residue and asparagine or the acetamide bond in
N-acetylhexosamines).
Lectin – A protein that specifically recognizes
and binds to glycans without catalyzing a modification
of the glycan.
Monosaccharide – A carbohydrate that cannot
be hydrolyzed into a simpler carbohydrate. It is
the building block of oligosaccharides and polysaccharides.
N-linked Glycan – Glycan covalently linked to
an asparagine residue of a polypeptide chain in
the consensus sequence: -Asn-X-Ser/Thr.
Non-reducing terminus – Outermost end of an
oligosaccharide or polysaccharide chain, which is
opposite to that of the reducing end.
O-linked Glycans – A glycan glycosidically
linked to the hydroxyl group of the amino acids
serine or threonine in the consensus GalNAcα1-
O-Ser/Thr.
Oligosaccharide – Linear or branched chain of
monosaccharides attached to one another via glycosidic
linkages. The number of monosaccharide
units can vary.
Polysaccharide – Glycan composed of repeating
monosaccharides, generally greater than ten
monosaccharide units in length.
Proteoglycan – Any protein with one or more
covalently attached glycosaminoglycan chains.
References:
- Spiro, R.G. (2002) Glycobiology, 12, 43R–56R.
- Bielik, A.M. and Zaia, J. (2010) Methods Mol. Biol., 600, 9–30.
- Kakehi, K. et al. (1994) J. Chromatogr A., 680, 209–215.
- Royle, L. et al. (2002) Anal. Biochem., 304, 70–90.
- Maley, F. et al. (1989) Anal. Biochem., 180, 195–204
- Beck, A. et al. (2008) Curr. Pharm. Biotechnol., 9, 482–501.
- Presta, L.G. (2008) Curr. Opin. Immunol., 20, 460–70.
- Iwai, T. et al. (2005) Natl. Acad. Sci. USA., 102, 4572–7.
- Iwai, T. et. al. (2002) J. Bio. Chem., 277, 12802–9.
- Capon, C. et al. (2001) Biochemical Journal, 358, 657–64.
- Springer, G.F. et al. (1997) Journal of Molecular Medicine, 75, 594–602.

