Plant and Insect Glycan Analysis
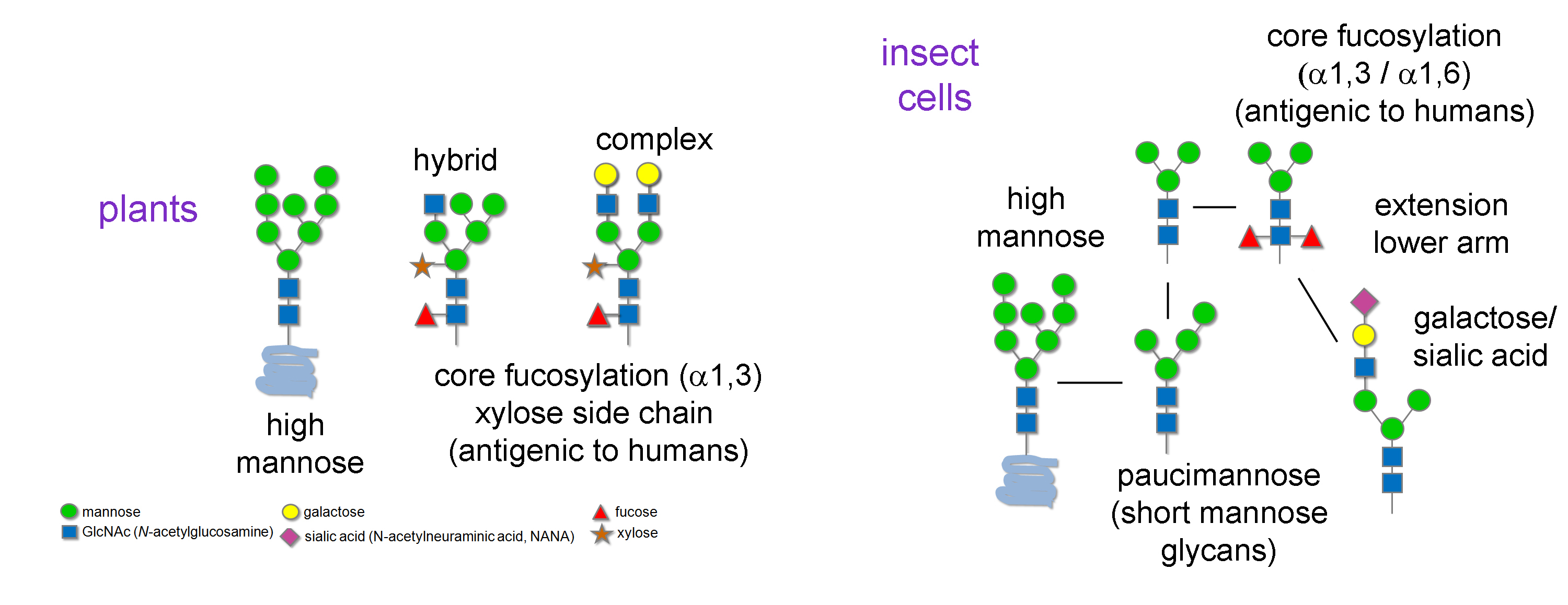
Return to Glycoprotein AnalysisUnless genetically engineered, plant N-glycans can be oligomannose, complex, hybrid and paucimannose with the N-glycans often modified at the core b-mannose with a b1-2 xylose residue. Additionally, the GlcNAc core of the N-glycan can be modified by an a1-3 fucose (Fig. 1). This core modification:
- Is known to be allergenic in humans
- Makes N-glycans resistant to PNGase F cleavage
- Can be cleaved by PNGase A and Endo D
Generally, insect N-glycans tend to be either high-mannose or paucimannose structures and unless genetically engineered the N-glycans also can be modified at the GlcNAc core by an a1-3 fucose (Fig. 1).
Figure 1: Typical N-glycan structures produced in plant and insects.
FAQs:
- What is the difference between PNGase F and PNGase A?
- Can PNGase A be used under non-denaturing (native) conditions?
- Which high mannose structures can PNGase A cleave?
- Can PNGase A cleave large, complex oligosaccharides?
- What happens to the asparagine after PNGase A removes the sugar?
- What is a good PNGase A substrate?
- Is PNGase A compatible with downstream analysis such as HPLC and Mass Spectrometry?
Videos
-
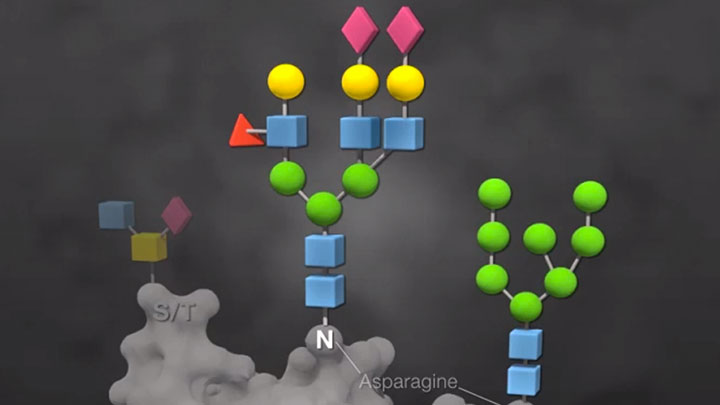
Overview of Glycobiology
Learn about the core sequences and common modifications of N-linked and O-linked glycans in this video. Analysis of these glycans can be accomplished with the use of deglycosylation enzymes, which can provide complete sugar removal with no protein degradation.
-

Behind the Paper: An engineered Fbs1 carbohydrate binding protein for selective capture of N-glycans and N-glycopeptides
Minyong and Jim summarize their recent Nature Communications publication describing selective capture of N-glycans and N-glycopeptides by an engineered high affinity Fbs1 carbohydrate binding protein.

