Mutate with Confidence! Tips and tools for successful site-directed mutagenesis experiments
Posted on Monday, June 26, 2023
By
Topic: Tips for the lab
If you're diving into site-directed mutagenesis experiments for the first time, or if you've been doing them for years but are looking to update your workflow, you may not be aware of various tools available to simplify the process and enhance your efficiency. From kits and enzyme mixes to web tools and streamlined protocols, we have the answers that will have you acing your site-directed mutagenesis experiments from start to finish!
Site-directed mutagenesis allows scientists to precisely modify discrete DNA sequences with insertions, deletions or substitutions. This ability to intentionally introduce targeted alterations at specific sites unlocks potential in a wide range of studies—from elucidating protein structure-function relationships, to enhancing enzymatic functionalities, to screening mutations for desired properties, or investigating disease mechanisms. Site-directed mutagenesis is also useful for domestication of plasmids with its ability to introduce or remove restriction endonuclease sites upstream of cloning experiments.
Over time, site-directed mutagenesis methodologies have evolved from complex and time-consuming multi-day experiments into more efficient and streamlined workflows. Keep reading to avoid wasting valuable time in the laboratory, and maybe even impress your labmates with the ease at which you generate your site-directed mutations!
While different approaches can be used to achieve the desired mutagenesis result, for this post, we will focus on site-directed mutagenesis as an in vitro procedure that uses custom-designed back-to-back primers to confer a desired mutation in a double-stranded DNA plasmid.
1. Start with effective primer design strategies for substitution, insertion or deletion
As with all experimental procedures, it is vital to take the time to plan mutagenesis experiments carefully. Arguably, the most significant contributing factor to successful site-directed mutagenesis reactions happens before you even pick up your pipette: primer design.
Many details need to be considered when designing primers for site-directed mutagenesis, including the incorporation of the desired mutation in the final product, as well as compatibility with intended downstream applications, such as cloning or sequencing.
Exponential amplification with back-to-back primers
You have the choice to use overlapping or back-to-back primers. First, let's clarify the concept of back-to-back primers: The 5´ ends are positioned adjacent to one another, with the primers facing opposite directions. To visualize this, imagine a clock face where both primers start at 12 o'clock—one moves clockwise while the other moves counterclockwise.
Using back-to-back primers offers distinct advantages in terms of input amount and flexibility in making desired modifications compared to using overlapping primers. Due to their length restriction, the use of overlapping primers imposes limitations on the number of base changes that can be incorporated within a single primer set. However, with back-to-back primers, inserting even up to 100 base pairs is feasible, as the bases to be inserted are added to the primer tails (see Figure 1, top right). Furthermore, the back-to-back primer strategy enables the deletion of any desired size by positioning the primers outside the target region, regardless of whether it spans a few base pairs or extends to hundreds of base pairs (see Figure 1, top middle). The back-to-back primer design results in a linear double-stranded PCR product.
Another advantage of using back-to-back primers is that amplification proceeds exponentially, and less input material is needed to achieve an adequate amount of the mutation-containing plasmid. In contrast, when overlapping primers are used, amplification is linear. It is important to note, though, that any polymerase errors that occur during PCR amplification will also propagate exponentially. Therefore, it is essential to use a high-fidelity polymerase to minimize errors. The NEB Q5® Site-Directed Mutagenesis Kit (NEB #E0554) uses back-to-back primers and incorporates Q5 High-Fidelity DNA Polymerase (NEB #M0491). Watch our video overview of the Q5 Site-Directed Mutagenesis Kit for more information.
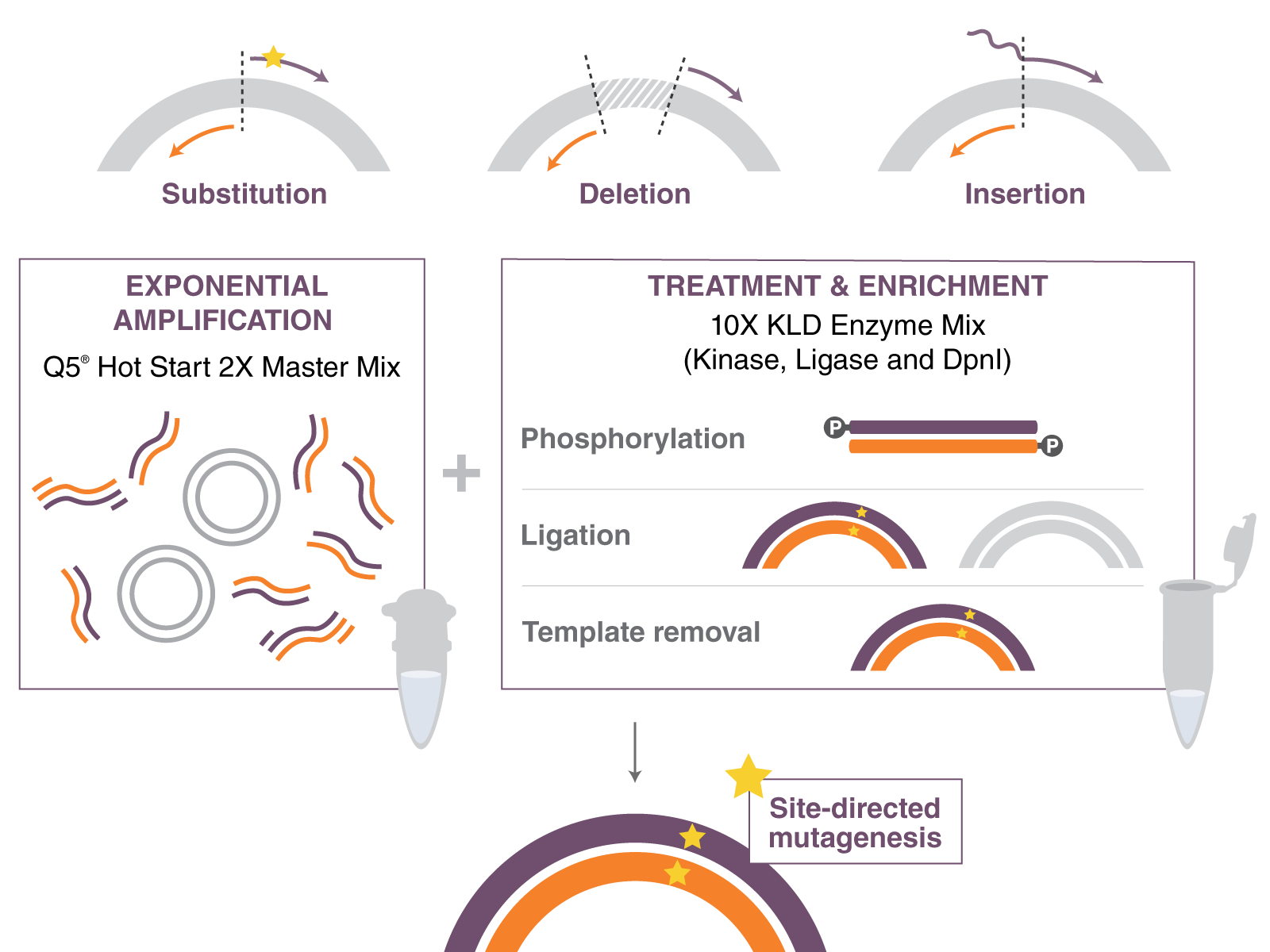
Figure 1: Top left: Substitutions are created by incorporating the desired nucleotide change(s) in the forward primer. The reverse primer is designed so that the 5´ ends of the two primers anneal back-to-back. Top Middle: Deletions are engineered by designing standard, non-mutagenic forward and reverse primers that flank the region to be deleted. Top right: Insertions less than or equal to 6 nucleotides are incorporated into the 5´ end of the forward primer, and the reverse primer anneals back-to-back with the forward primer. Larger insertions are created by incorporating half of the desired insertion into the 5´ ends of both primers.
The first step is an exponential amplification using primers and a master mix formulation of Q5 Hot Start High-Fidelity DNA Polymerase. The second step involves incubation with a unique enzyme mix containing a kinase, a ligase and DpnI. Together, these enzymes allow for rapid circularization of the PCR product and removal of the template DNA. The last step is a high-efficiency transformation into competent cells.
The NEBaseChanger web tool has new features for primer design
New England Biolabs® scientists and bioinformaticians built the NEBaseChanger® web tool to remove much of the uncertainty, and potential for errors, out of primer design. This open access software allows you to input a plasmid sequence and select the mutagenesis type (substitution, insertion, deletion) to generate primer sequences and recommended annealing temperature (taking into account the use of Q5 High-fidelity DNA Polymerase).
We have updated this popular web tool, introducing new features to enhance its functionality and cater to the evolving needs of scientists and researchers in their primer design processes (Figure 2). Users can now take advantage of the batch input option, which allows for the simultaneous generation of multiple primer sets. It is also possible to opt for mutation sequences in the primer tails. Moreover, the updated tool has improved compatibility with standard mixed or ambiguous base entries and a new option to perform substitutions based on either nucleotide or amino acid positions.
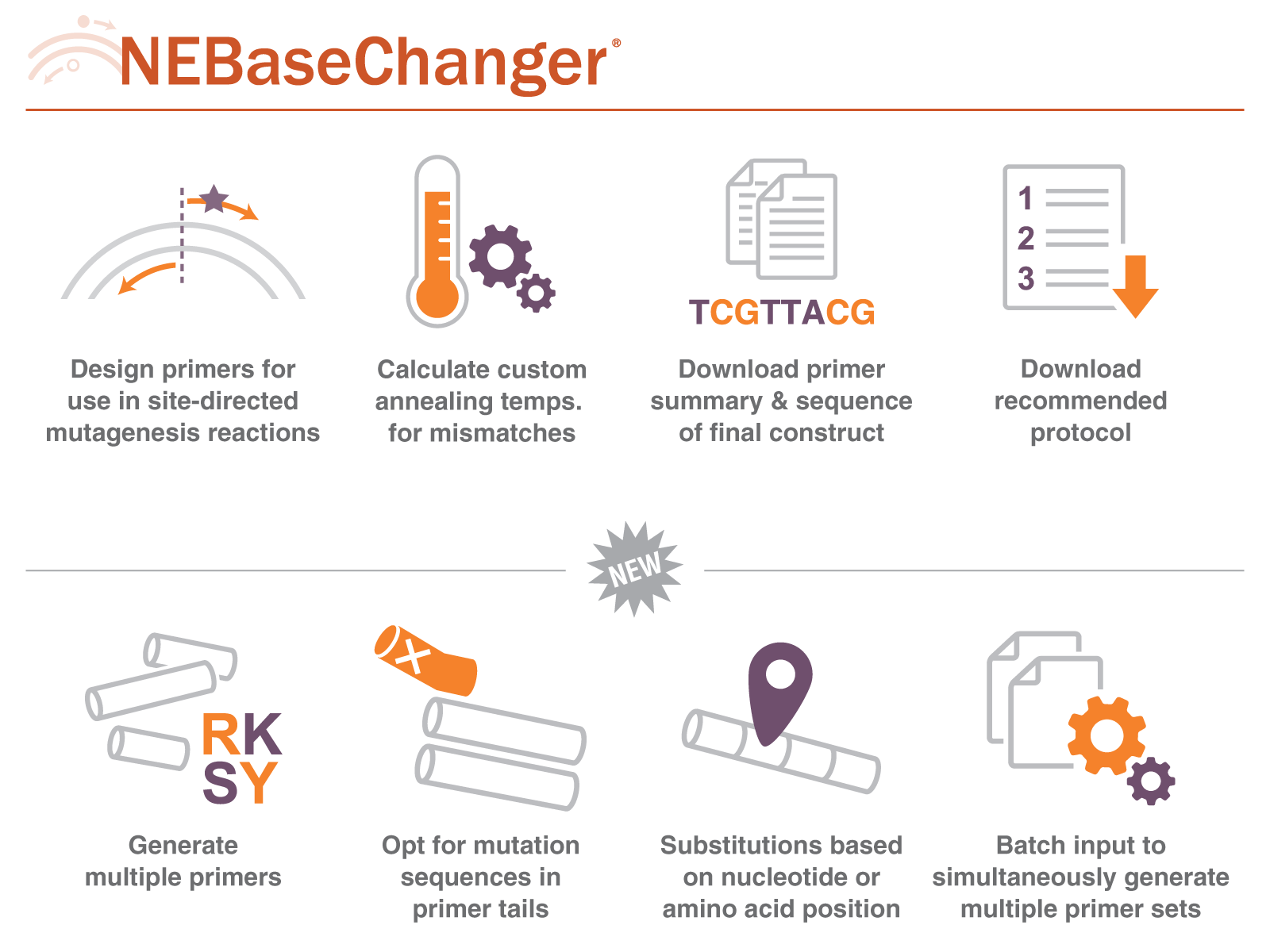
Figure 2: A summary of the features of the NEBaseChanger, our web tool that assists you in primer design for your mutagenesis experiments.
2. Ensure amplification with optimized PCR conditions
You're off to a great start once you have designed the mutagenic primers using NEBaseChanger. This takes one of the major pitfalls of site-directed mutagenesis off the table.
The following tips can also help you optimize your PCR reaction:
- Anneal – a higher annealing temperature is required if using a high-fidelity DNA polymerase. This is already taken into account if using NEBaseChanger to design your primers, but if not, we recommend a Tm+3 rule: that is, add 3°C to the Tm recommended by your oligo supplier.
Extend – avoid long extension times. Use an extension time of 20-30 seconds per kilobase, although this can vary for very large transcripts.
Denature – use a denaturation temperature of 98°C. - Use only 1 unit of Q5 DNA Polymerase per 25 µl reaction.
- Use a final primer concentration of 0.5 µM each.
- Run your PCR-amplified fragment on a gel before using it. While this is not required, it is helpful to run the linearized PCR product on an agarose gel to establish you have one single PCR product represented by a clean band. If, for example, you find you have more than one band or a smear, more optimization is required.
3. Recircularize your PCR product and remove wildtype background all at once
The non-mutated template is usually removed following the PCR reaction using the restriction enzyme DpnI because it selectively digests methylated DNA. For example, DpnI digests plasmids that are propagated and isolated from E. coli and are, therefore, methylated; but PCR products do not contain methyl groups and will not be digested by DpnI.
The KLD enzyme mix (NEB #M0554) combines three enzymes: a kinase, a ligase and DpnI. The kinase phosphorylates the 5´ end of the linearized plasmid, the ligase recircularizes it, and the DpnI (as mentioned above) digests the wildtype template, leaving behind the PCR product containing the desired mutation. The reaction time is as short as 5 minutes. This convenient three-enzyme mix is supplied with the Q5 Site-Directed Mutagenesis Kit.
If you have high background following transformation (i.e., many colonies that still contain the wildtype plasmid), the background can be further reduced by increasing the KLD incubation time to 30-60 minutes.
4. Secure the target clone with efficient transformation and screening
The potential mutation-carrying plasmid can be transformed into most competent E. coli strains. The efficiency of transformation will vary depending on factors such as the plasmid size and the quality and effectiveness of the competent cells utilized. This brings us to another common query—whether or not to use homebrew competent cells.
Competent E. coli cells: to homebrew or not?
The decision to use homebrew competent E. coli cells for plasmid transformation depends on several factors. Homebrew competent cells can be prepared in the laboratory using specific protocols, offering potential cost savings compared to commercially available competent cells. However, it requires specialized knowledge and can be time-consuming. Inadequate preparation can lead to low transformation efficiency or unreliable results. Commercially available competent cells are optimized for high efficiency and reproducibility and undergo rigorous quality control to ensure consistent performance. While homebrew cells may be cheaper initially, factoring in the cost of labor, reagents, and potential variability in results is crucial for a comprehensive cost-benefit analysis.
The Q5 Site-Directed Mutagenesis kit is supplied with or without 5-alpha competent E. coli cells. These cells support transformation with plasmids up to at least 20 kb in length.
Why am I getting wildtype colonies?
The most common problem by far that our technical help team hears is when customers get wildtype colonies instead of colonies with their desired mutation. Often, this is because the PCR reaction needs to be optimized. For example, if the primers don’t bind well or bind in a different location than the intended location, they can create the wrong amplicon or no amplicon at all. If nothing is amplified, then the only transformable component of the reaction is some residual wildtype plasmid.
- We recommend using ≤ 10 ng of template in the PCR step.
- You can reduce the background by increasing the KLD incubation time to 30-60 minutes.
- Ensure proper design of the mutagenic primers, using our online tool NEBaseChanger
- Optimize PCR conditions. High-fidelity polymerases benefit from a Tm+3 annealing temperature as an estimate.
It should be noted that one feature that the NEBaseChanger web tool does not have is the ability to check the specificity of the primers against the whole plasmid template. While the tool provides optimal primer design for your desired mutation, it cannot predict if that primer pair might also bind somewhere else on the plasmid. So, checking your primers against your entire template before ordering them is crucial.
Site-directed Mutagenesis Troubleshooting
Take your time to plan your experiment carefully. Follow these tips and utilize our videos and web tool. This will minimize the chances of pulling an agar plate from the warm room only to discover no colonies on it!
And probably the most valuable tip of all – Call our Technical Support Team sooner rather than later. Our team is here to help.
New England Biolabs offers a suite of reagents, protocols and helpful application notes for Site Directed Mutagenesis workflows.
If you’re planning to do multi-site mutagenesis, take a look at our recent Application Note, Improved methods for site-directed mutagenesis using NEBuilder® HiFi DNA Assembly Master Mix (NEB #E2621) and utilize our updated NEBaseChanger webtool that now designs primers for multi site-directed mutagenesis with NEBuilder HiFi.
NEB will not rent, sell or otherwise transfer your data to a third party for monetary consideration. See our Privacy Policy for details. View our Community Guidelines.
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. See www.neb.com/trademarks. The use of these products may require you to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
Don’t miss out on our latest NEBinspired blog releases!
- Sign up to receive our e-newsletter
- Download your favorite feed reader and subscribe to our RSS feed
Be a part of NEBinspired! Submit your idea to have it featured in our blog.


