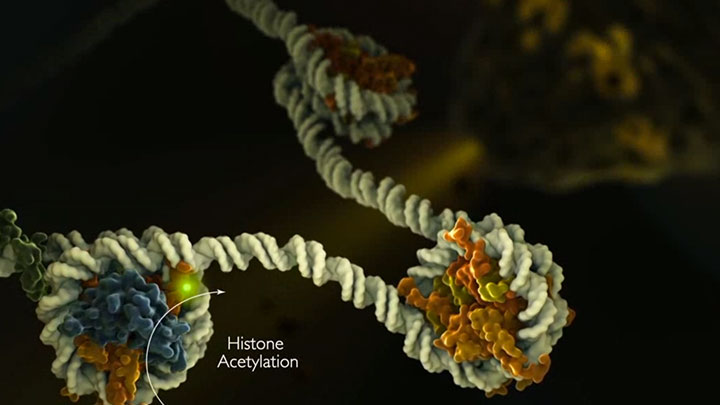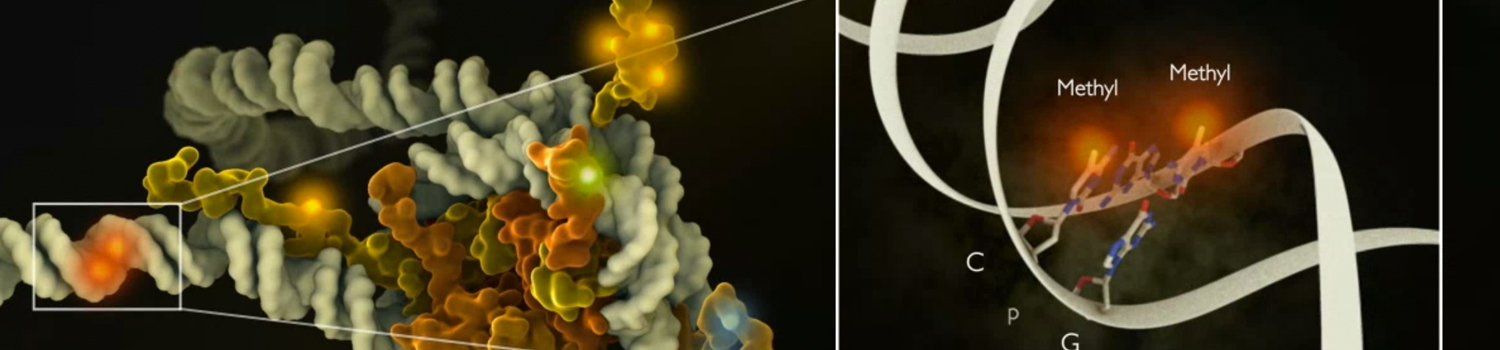
Epigenetics
Choose Type:
-
Epigenetics - Expanding on Genomic Foundations
-
Enzymatic Methyl-seq: The Next Generation of Methylome Analysis
- Epigenetics Brochure
- NicE-seq: High Resolution Open Chromatin Profiling (2018)
- EM-seq™ Enables Accurate and Precise Methylome Analysis of Challenging DNA Samples (2019)
- Enzymatic Methyl-seq: Next Generation Methylomes (2019)
- Uncovering the Cannabis sativa Methylome Through Enzymatic Methyl-seq (2019)
- EM-seq™ enables accurate and robust methylation detection of cell free DNA and FFPE DNA sample types
Feature Articles
Brochures
Posters
- Woodcock, C.B.l, Yu, D., Hajian, T., Li, J., Huang, Y., Dai, N., Corrêa, I.R., Jr., Wu, T., Vedadi, M., Zhang, X., Cheng, X (2019) Human MettL3 MettL14 complex is a sequence specific DNA adenine methyltransferase active on single strand and unpaired DNA in vitro Cell Discov ; 5:63, PubMedID: 31885874, DOI: 10.1038/s41421-019-0136-4
- Murray, Iain; Morgan, Rick; Luyten, Yvette; Fomenkov, Alexey; Correa, Ivan; Dai, Nan; Roberts, Rich; (2018) The non-specific adenine DNA methyltransferase M.EcoGII. Nucleic Acids Res; (2), 840-848. PubMedID: 29228259, DOI: 10.1093/nar/gkx1191
- Mooijman D, Dey S S, Boisset JC, Crosetto N, van Oudenaarden A (2016) Single-cell 5hmC sequencing reveals chromosome-wide cell-to-cell variability and enables lineage reconstruction Nat Biotechnol; 34, 852-857. PubMedID: 27347753, DOI: 10.1038/nbt.3598
- Marx V. (2016) Genetics: profiling DNA methylation and beyond Nat Methods; 13, 119-122. DOI: 10.1038/nmeth.3736
- Page A., Paoli P., Salvador E., White S., French J., Mann J. (2015) Hepatic Stellate Cell Transdifferentiation Involves Genome-Wide Remodeling of the DNA Methylation Landscape J Hepatol; PubMedID: 26632634, DOI: 10.1016/j.jhep.2015.11.024
- Pirone-Davies, C., Hoffmann, M., Roberts, R.J., Muruvanda, T., Timme, R.E., Strain, E., Luo, Y., Payne, J., Luong, K., Song, Y., Tsai, Y.C., Boitano, M., Clark, T.A., Korlach, J., Evans, P.S., Allard, M.W. (2015) Genome-Wide Methylation Patterns in Salmonella enterica Subsp. enterica Serovars PLoS One; 10(4), e0123639. PubMedID: 25860355
- Pais, J.E., Dai, N., Tamanaha, E., Vaisvila, R., Fomenkov, A.I., Bitinaite, J., Sun, Z., Guan, S., Correa, I.R. Jr., Noren, C.J., Cheng, X., Roberts, R.J., Zheng, Y. and Saleh, L. (2015) Biochemical characterization of a Naegleria TET-like oxygenase and its application in single molecule sequencing of 5-methylcytosine Proc Natl Acad Sci U S A; 112(14), 4315-4321. PubMedID: 25831492
- Estève P., Zhang G., Ponnaluri V., Kanneganti (2015) Binding of 14-3-3 reader proteins to phosphorylated DNMT1 facilitates Nucleic Acids Res; PubMedID: 26553800, DOI: 10.1093/nar/gkv1162
- Wee E., Ngo T., Trau M. (2015) A simple bridging flocculation assay for rapid, sensitive and stringent detection of gene specific DNA methylation Sci Rep; 5, 15028. PubMedID: 26458746, DOI: 10.1038/srep15028
- Chernov AV., Reyes L., Peterson S., Strongin AY. (2015) Depletion of CG-Specific Methylation in Mycoplasma hyorhinis Genomic DNA after Host Cell Invasion PLoS One; 10, e0142529. PubMedID: 26544880, DOI: 10.1371/journal.pone.0142529
- Kienhöfer S., Musheev M., Stapf U., Helm M., Schomacher L., Niehrs C., Schäfer A. (2015) GADD45a physically and functionally interacts with TET1Publication Differentiation; PubMedID: 26546041, DOI: 10.1016/j.diff.2015.10.003
- Hughes J.R., Roberts N., McGowan S., Hay D., Giannoulatou E., Lynch M., De Gobbi M., Taylor S., Gibbons R., Higgs D.R. (2014) Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment Nat Genet; 46 (2), 205-212. PubMedID: 24413732, DOI: doi:10.1038/ng.2871
- Chandra S, Baribault C, Lacey M, Ehrlich M (2014) Myogenic differential methylation: diverse associations with chromatin structure Biology (Basel); 3(2), 426-51. PubMedID: 24949935, DOI: 10.3390/biology3020426
- Sexton T, Kurukuti S, Mitchell JA, Umlauf D, Nagano T, Fraser P (2012) Sensitive detection of chromatin coassociations using enhanced chromosome conformation capture on chip Nat Protoc; 7(7), 1335-50. PubMedID: 22722369, DOI: 10.1038/nprot.2012.071
- Wolff, E.M. et al. (2011) Hypomethylation of a LINE-1 Promoter Activates an Alternate Transcript of the MET Oncogene in Bladders with Cancer PLoS Genet; 6, 4:e1000917 . PubMedID: 20421991
- Lieberman-Aiden E., van Berkum N., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B., Sabo P., Dorschner M., Sandstrom R., Bernstein B., BenderM. A., Groudine M., Gnirke A., Stamatoyannopoulos J., Mirny L., Lander E., Dekker J. (2009) Comprehensive mapping of long range interactions reveals folding principles of the human genome Science; Oct 9; 326(5950) , 289–293.
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.