Detailed Characterization of Several Glycosidase Enzymes
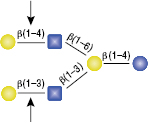
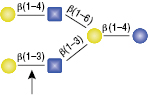
β1-3 Galactosidase
The GlcNac (β1-6) residue is the only anomeric configuration that can effect the specificity of the enzyme enabling cleavage of the non-reducing β1-4 Galactose.
Selling concentration of the enzyme will cut the β1-4 Galactose linkage as shown in (A) due to the adjacent GlcNAcβ1-6 anomer. This cleavage will not occur if the selling concentration of the enzyme is diluted 16-fold, shown in (B).
Chart Legend: Gal Glc
Glc Man
Man GalNAc
GalNAc GlcNAc
GlcNAc Fuc
Fuc NeuAc
NeuAc R = any sugar
R = any sugar
β1-4 Galactosidase
Reactions (A), (B) and (C) contained 2 units, 4 units and 8 units of β1-4 Galactosidase, respectively, and either 1X G4 or 1X G6 reaction buffer in a total reaction volume of 10 µl. Reactions were incubated at 37°C.
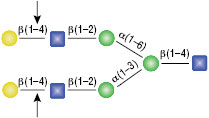
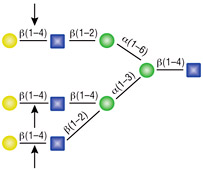
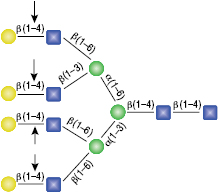
β-N-Acetylglucosaminidase
All reactions contained 4 units of β-N-Acetylglucosaminidase, 1X G1 reaction buffer and 1X BSA in a total reaction volume of 10 µl. Reactions (B), (C) and (D) were treated with 8 units of β1-4 Galactosidase prior to treatment with β-N-Acetylglucosaminidase. Reactions were incubated at 37°C.
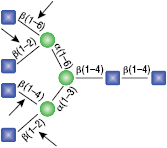
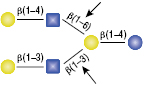
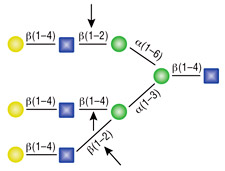
α1-2,3 Mannosidase
All reactions contained 32 units of α1-2,3 Mannosidase, 1X G6 reaction buffer and 1X BSA in a total reaction volume of 10 µl. Reactions were incubated at 37°C.
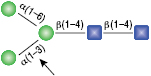
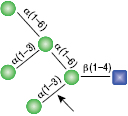
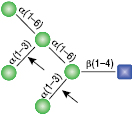
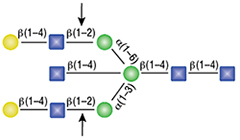
α1-6 Mannosidase
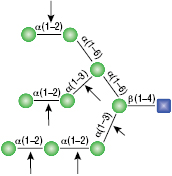
All reactions contained 32 units of α1-2,3 Mannosidase (NEB #P0729), 40 units of α1-6 Mannosidase, 1X G6 reaction buffer and 1X BSA in a total reaction volume of 10 µl. Reactions were incubated at 37°C. The substrate depicted in (E) will not cut to completion. If this structure exists in any substrate it will be impervious to cleavage by α1-6 Mannosidase. Note: When used alone, α1-6 Mannosidase will still act only on linear substrates. When used in conjunction with α1-2,3 Mannosidase, the α1-6 Mannosidase will cleave α1-6 Mannose residues from branched carbohydrate substrates.
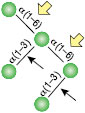
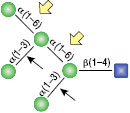
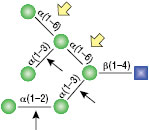
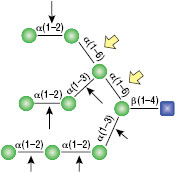
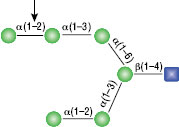
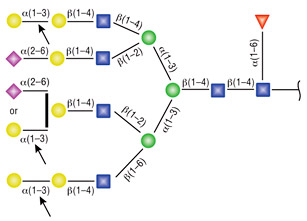
α1-3,6 Galactosidase
Reaction (A) contained 24 units of α1-3,6 Galatosidase and 100 units of Neuraminidase (NEB #P0720), followed by a heat kill at 65°C for 10 minutes and a 2 hour digestion with 16 units of β1-4 Galactosidase (NEB #P0730).
The reaction in (B) contained 4 units of α1-3, 6 Galactosidase, 1X G6 reaction buffer and 1X BSA in a total reaction volume of 20 µl.
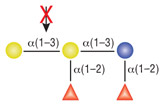
α1-2 Fucosidase
Reactions (A), (B) and (D) contained 1X G4 reaction buffer and reaction (C) contained 1X G6 reaction buffer. All reactions contain 1X BSA in a total reaction volume of 10 µl and all reactions were incubated at 37°C. All reactions contained 20 units of α1-2 Fucosidase. Reaction (C) also contained 20 units of α-N-Acetylgalactosaminidase (NEB #P0734). In reaction (C), the branched α1-2 fucose is removed in the presence of both enzymes, but not by α1-2 Fucosidase alone.
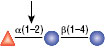
PNGase F
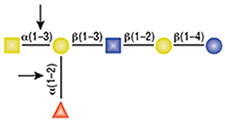
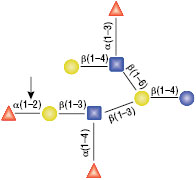
PNGase F is not able to cleave N-linked glycans from glycoproteins when the innermost GlcNAc residue is linked to an α1-3 Fucose residue (1). This modification is most commonly found in plant and some insect glycoproteins.
(1) Tretter, V. et al. (1991) Eur. J. Biochem. 199, 647–652. PMID: 1868849
Remove-iT™ Endo S
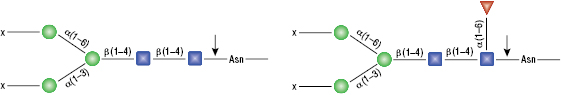
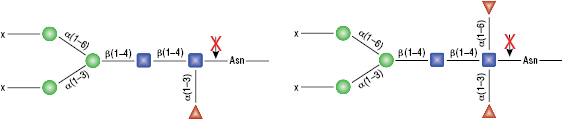
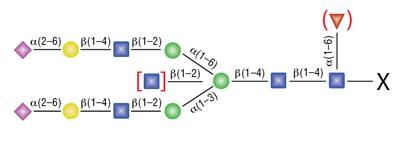
Remove-iT Endo S does not have a strict peptide requirement for activity, thus the “X” can be a protein, peptide, Asparagine, or free glycan. Remove-iT Endo S is active on a substrate with or without core a(1-6)fucosylation as well as with or without a bisecting N-acetylglucosamine. Triantennary and tetrantennary sialyted or asialo glycans are not a substrate for Remove-iT Endo S.
Remove-iT™ Endo D
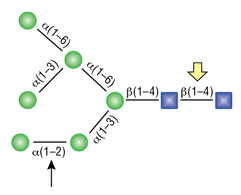
Remove-iT Endo D, also known as Endoglycosidase D, is a recombinant glycosidase, which cleaves within the chitobiose core of paucimannose N-linked glycans, with or without extensions in the antennae. Remove-iT Endo D is tagged with a chitin binding domain (CBD) for easy removal from a reaction, and is supplied glycerol-free for optimal performance in HPLC- and MS- intensive methods.
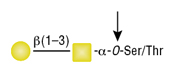
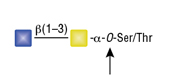
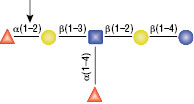
O-Glycosidase (Endo-a-N-Acetylgalactosaminidase)
Reactions (A) and (B) contained 40,000 units of Endo-a-N-Acetylgalactosaminidase, 100 units of Neuraminidase and 1X G7 reaction buffer in a total reaction volume of 100 μl. The Reactions were incubated at 37°C and released O-linked disaccharide carbohydrates were determined by Morgan and Elson Assay.