Protocol for use with NEBNext® Poly(A) mRNA Magnetic Isolation Module (E7490) and NEBNext® Ultra™ II RNA Library Prep Kit for Illumina® (E7770, E7775)
 |
This is a point where you can safely stop the protocol and store the samples prior to proceeding to the next step in the protocol. |
 |
This caution sign signifies a step in the protocol that has two paths leading to the same end point but is dependent on a user variable, like the type of RNA input. |
 |
Colored bullets indicate the cap color of the reagent to be added |
The protocol has been optimized using high quality Universal Human Reference Total RNA.
RNA Sample Recommendations
RNA Integrity:
Assess the quality of the Input RNA by running the RNA sample on an Agilent Bioanalyzer® RNA 6000 Nano/Pico Chip. For PolyA mRNA enrichment, high quality RNA with a RIN score > 7 is required.
RNA Sample Requirements:
The RNA sample should be free of salts (e.g. Mg2+, or guanidinium salts, divalent cation chelating agents (e.g. EDTA or EGTA) or organics (e.g. phenol or ethanol). RNA must be free of DNA. gDNA is a common contaminant from RNA preps. It may be carried over from the interphase of organic extractions or when the silica matrix of solid phase RNA purification methods is overloaded. If the total RNA sample may contain gDNA contamination, treat the sample with DNase I to remove all traces of DNA (DNase is not provided in this kit). After treatment with DNase I the enzyme should be removed from the sample. Any residual activity of the DNase I may degrade the oligos necessary for the enrichment. DNase I can be removed from the extraction using phenol/chloroform extraction and ethanol precipitation.
Input Amount Requirement:
10 ng–1 μg DNA-free total RNA quantified by Qubit® Fluorometer and quality checked by Bioanalyzer. The protocol is optimized for approximately 200 bp RNA inserts. To generate libraries with longer RNA insert sizes, refer to Appendix A (Section 6) for recommended fragmentation times and size selection conditions.
Keep all the buffers on ice, unless otherwise indicated.
1.1.  Preparation of First Strand Reaction Buffer and Random Primer Mix
Preparation of First Strand Reaction Buffer and Random Primer Mix
1.1.1. Prepare the First Strand Synthesis Reaction Buffer and Random Primer Mix (2X) in a nuclease-free microcentrifuge tube as follows:
| COMPONENT | VOLUME |
|---|---|
 (lilac) NEBNext First Strand Synthesis Reaction Buffer (lilac) NEBNext First Strand Synthesis Reaction Buffer |
8 µl |
 (lilac) NEBNext Random Primers (lilac) NEBNext Random Primers |
2 µl |
| Nuclease-free water |
10 µl |
| Total Volume |
20 µl |
1.1.2. Mix thoroughly by pipetting up and down ten times.
Note: Keep the mix on ice until mRNA is purified. It will be used in Step 1.2.36 and 1.2.39.
1.2. mRNA Isolation, Fragmentation and Priming Starting with Total RNA
1.2.1. Dilute the total RNA with nuclease-free water to a final volume of 50 μl in a nuclease-free 0.2 ml PCR tube and keep on ice.
1.2.2. To wash the Oligo dT Beads, add the following to a 1.5 ml nuclease-free tube. If preparing multiple libraries, beads for up to 10 samples can be added to a single 1.5 ml tube for subsequent washes (use magnet NEB #S1506 for 1.5 ml tubes). The purpose of this step is to bring the beads from the storage buffer into the binding buffer. The 2X Binding Buffer does not have to be diluted
for this step.
| COMPONENT | VOLUME |
|---|---|
| Oligo dT Beads d(T)25 |
20 µl |
| RNA Binding Buffer (2X) |
100 µl |
| Total Volume |
120 µl |
1.2.3. Wash the beads by pipetting up and down six times.
1.2.4. Place the tube on the magnet and incubate at room temperature until the solution is clear (~2 minutes).
1.2.5. Remove and discard all of the supernatant from the tube. Take care not to disturb the beads.
1.2.6. Remove the tube from the magnetic rack.
1.2.7. Add 100 μl RNA Binding Buffer (2X) to the beads and wash by pipetting up and down six times. If preparing multiple libraries, add 100 µl RNA Binding Buffer (2X) per sample. The Binding Buffer does not have to be diluted.
1.2.8. Place the tubes on the magnet and incubate at room temperature until the solution is clear (~2 minutes).
1.2.9. Remove and discard the supernatant from the tube. Take care not to disturb the beads.
1.2.10. Add 50 μl RNA Binding Buffer (2X) to the beads and mix by pipetting up and down until beads are homogenous. If preparing multiple libraries, add 50 μl RNA Binding Buffer (2X) per sample. This first binding step removes most of the non target RNA.
1.2.11. Add 50 μl beads to each RNA sample from Step 1.2.1. Mix thoroughly by pipetting up and down six times.
1.2.12. Place the tube in a thermocycler and close the lid. Heat the sample at 65°C for 5 minutes and cool to 4°C with the heated lid set at ≥ 75°C to denature the RNA and facilitate binding of the mRNA to the beads.
1.2.13. Remove the tube from the thermocycler when the temperature reaches 4°C.
1.2.14. Mix thoroughly by pipetting up and down six times. Place the tube on the bench and incubate at room temperature for 5 minutes to allow the mRNA to bind to the beads.
1.2.15. Place the tube on the magnetic rack at room temperature until the solution is clear (~2 minutes).
1.2.16. Remove and discard all of the supernatant. Take care not to disturb the beads.
1.2.17. Remove the tube from the magnetic rack.
1.2.18. Wash the beads by adding 200 μl of Wash Buffer to the tube to remove unbound RNA. Gently pipette the entire volume up and down 6 times to mix thoroughly.
1.2.19. Place the tube on the magnetic rack at room temperature until the solution is clear (~2 minutes).
1.2.20. Remove and discard all of the supernatant from the tube. Take care not to disturb the beads.
1.2.21. Remove the tube from the magnetic rack.
1.2.22. Repeat steps 1.2.18–1.2.21.
1.2.23. Add 50 μl of Tris Buffer (provided in NEB #E7490 kit) to each tube. Gently pipette up and down 6 times to mix thoroughly.
1.2.24. Place the tube on the thermocycler. Close the lid and heat the samples at 80°C for 2 minutes, then cool to 25°C with the heated lid set at ≥ 90°C to do the first elution of the mRNA from the beads.
1.2.25. Remove the tube from the thermocycler when the temperature reaches 25°C.
1.2.26. Add 50 μl of RNA Binding Buffer (2X) to the sample to allow the mRNA to re-bind to the beads. Mix thoroughly by gently pipetting up and down six times.
1.2.27. Incubate the tube at room temperature for 5 minutes.
1.2.28. Place the tube on the magnetic rack at room temperature until the solution is clear (~2 minutes).
1.2.29. Remove and discard the supernatant from the tube. Take care not to disturb the beads.
1.2.30. Remove the tube from the magnetic rack.
1.2.31. Wash the beads by adding 200 μl of Wash Buffer. Gently pipette the entire volume up and down 6 times to mix thoroughly.
1.2.32. Spin down the tube briefly to collect the liquid from the wall and lid of the tube.
Note: It is important to spin down the tube to prevent carryover of the Wash Buffer in subsequent steps.
1.2.33 Place the tube on the magnet at room temperature until the solution is clear (~2 minutes).
1.2.34. Remove and discard all of the supernatant from the tube. Take care not to disturb the beads that contains the mRNA.
Note: It is important to remove all of the supernatant to successfully fragment the mRNA in the subsequent steps. Spin down the tube. Place the tube on the magnetic rack and with a 10 μl tip, remove all of the wash buffer. (Caution: Do not disturb beads that contain the mRNA). Avoid letting the beads dry out before adding elution buffer.
1.2.35. Remove the tube from the magnetic rack.
 |
The next step provides a fragmentation incubation time resulting in
an RNA insert size of ~ 200 nt. For RNA insert sizes > 200 nt, refer
to Section 6 (Appendix A) for recommended fragmentation times in Step
1.2.37. |
1.2.36. To elute the mRNA from the beads and fragment, add 11.5 μl of the First Strand Synthesis Reaction Buffer and Random Primer Mix (2X) prepared in Step 1.1.2, pipette up and down six times to resuspend the beads.
1.2.37 Incubate the sample in a thermal cycler with the heated lid set at 105°C as follows:
15 minutes at 94°C
Hold at 4°C*
*Immediately transfer the tube to ice for 1 minute as soon as it is cool enough to handle (~65°C)
1.2.39. Collect the fragmented mRNA by transferring 10 μl of the supernatant to a nuclease-free 0.2 ml PCR tube.
Note 1: If the supernatant volume recovered is less than 10 μl for any reason, bring the volume up to 10 μl by adding the First Strand Synthesis Reaction Buffer and Random Primer Mix (2X) prepared in Step 1.1.2 and continue with the protocol.
Note 2: Avoid transferring the magnetic beads.
1.2.40. Place the tube on ice and proceed directly to First Strand cDNA Synthesis.
1.3 First Strand cDNA Synthesis
1.3.1. Assemble the first strand cDNA synthesis reaction on ice by adding the following components into fragmented and primed RNA from Step 1.2.40.
| FIRST STRAND cDNA SYNTHESIS REACTION | VOLUME |
|---|---|
| Fragmented and primed RNA (Step 1.2.40) | 10 µl |
| Nuclease-free Water | 8 µl |
 (lilac) NEBNext First Strand Synthesis Enzyme Mix (lilac) NEBNext First Strand Synthesis Enzyme Mix |
2 µl |
| Total Volume |
20 µl |
1.3.2. Mix thoroughly by pipetting up and down at least 10 times.
1.3.3.
 Incubate the sample in a preheated thermocycler with the heated lid set at ≥ 80°C as follows:
Incubate the sample in a preheated thermocycler with the heated lid set at ≥ 80°C as follows:Note: If you are following recommendations in Appendix A, for longer RNA fragments (creating inserts > 200 bases), increase the incubation at 42°C from 15 minutes to 50 minutes at Step 2.
Step 1: 10 minutes at 25°C
Step 2: 15 minutes at 42°C
Step 3: 15 minutes at 70°C
Step 4: Hold at 4°C
1.4.Second Strand cDNA Synthesis
1.4.1. Assemble the second strand cDNA synthesis reaction on ice by adding the following components into the first strand synthesis reaction product from Step 1.3.4.
| SECOND STRAND SYNTHESIS REACTION | VOLUME |
|---|---|
| First-Strand Synthesis Product (Step 1.3.4) | 20 µl |
 (orange) NEBNext Second Strand Synthesis Reaction Buffer (10X) (orange) NEBNext Second Strand Synthesis Reaction Buffer (10X) |
8 µl |
 (orange) NEBNext Second Strand Synthesis Enzyme Mix (orange) NEBNext Second Strand Synthesis Enzyme Mix |
4 µl |
| Nuclease-free Water | 48 µl |
| Total Volume |
80
µl |
1.4.2. Keeping the tube on ice, mix thoroughly by pipetting the reaction up and down at least 10 times.
1.4.3. Incubate in a thermocycler for 1 hour at 16°C with the heated lid set at ≤ 40°C (or off).
1.5. Purification of Double-stranded cDNA using SPRIselect Beads or NEBNext Sample Purification Beads
1.5.1. Vortex SPRIselect Beads or NEBNext Sample Purification Beads to resuspend.
1.5.2. Add 144 μl (1.8X) of resuspended beads to the second strand synthesis reaction (~80 μl). Mix well on a vortex mixer or by pipetting up and down at least 10 times.
1.5.3. Incubate for at least 5 minutes at room temperature.
1.5.4. Briefly spin the tube in a microcentrifuge to collect any sample from the sides of the tube. Place the tube on a magnetic rack to separate beads from the supernatant. After the solution is clear, carefully remove and discard the supernatant. Be careful not to disturb the beads, which contain DNA. (Caution: do not discard beads).
1.5.5. Add 200 μl of freshly prepared 80% ethanol to the tube while in the magnetic stand. Incubate at room temperature for 30 seconds, and then carefully remove and discard the supernatant.
1.5.6. Repeat Step 1.5.5 once for a total of 2 washing steps.
1.5.7. Air dry the beads for 5 minutes while the tube is on the magnetic rack with the lid open.
Caution: Do not over-dry the beads. This may result in lower recovery of DNA target. Elute the samples when the beads are still dark brown and glossy looking, but when all visible liquid has evaporated. When the beads turn lighter brown and start to crack they are too dry.
1.5.8. Remove the tube from the magnet. Elute the DNA target from the beads by adding 53 μl 0.1X TE Buffer (provided) to the beads. Mix well on a vortex mixer or by pipetting up and down ten times. Briefly spin the tube and incubate for 2 minutes at room temperature. Place the tube on the magnetic rack until the solution is clear.
1.5.9. Remove 50 µl of the supernatant and transfer to a clean nuclease-free PCR tube.
| If you need to stop at this point in the protocol, samples can be stored at –20°C. |
1.6 End Prep of cDNA Library
1.6.1. Assemble the end prep reaction on ice by adding the following components to second strand synthesis product from Step 1.5.9.
| END PREP REACTION | VOLUME |
|---|---|
| Second Strand cDNA Synthesis Product (Step 1.5.9) |
50 µl |
 (green) NEBNext Ultra II End Prep Reaction Buffer (green) NEBNext Ultra II End Prep Reaction Buffer |
7 µl |
 (green) NEBNext Ultra II End Prep Enzyme Mix (green) NEBNext Ultra II End Prep Enzyme Mix |
3 µl |
| Total Volume |
60
µl |
1.6.2 Set a 100 μl or 200 μl pipette to 50 μl and then pipette the entire volume up and down at least 10 times to mix thoroughly. Perform a quick spin to collect all liquid from the sides of the tube.
Note: It is important to mix well. The presence of a small amount of bubbles will not interfere with performance.
1.6.3. Incubate the sample in a thermocycler with the heated lid set at ≥ 75°C as follows:
30 minutes at 20°C
30 minutes at 65°C
Hold at 4°C
1.7 Adaptor Ligation
1.7.1.
 Dilute the
Dilute the  (red) NEBNext Adaptor* prior to setting up the ligation reaction in ice-cold
Adaptor Dilution Buffer and keep the diluted adaptor on ice.
(red) NEBNext Adaptor* prior to setting up the ligation reaction in ice-cold
Adaptor Dilution Buffer and keep the diluted adaptor on ice.| TOTAL RNA input | DILUTION REQUIRED |
|---|---|
| 1,000 ng–250 ng | 5-fold dilution in Adaptor Dilution Buffer |
| 249 ng–100 ng |
25-fold dilution in Adaptor Dilution Buffer |
| 99 ng–10 ng | 100-fold dilution in Adaptor Dilution Buffer |
*The NEBNext adaptor is provided in NEBNext oligos kit. NEB has several oligo kit options, which are supplied separately from the library prep kit.
1.7.2. Assemble the ligation reaction on ice by adding the following components, in the order given, to the end prep reaction product from Step 1.6.4.
| LIGATION REACTION |
VOLUME |
|---|---|
| End Prepped DNA (Step 1.6.4) | 60 µl |
| Diluted Adaptor (Step 1.7.1) |
2.5 µl |
 (red) NEBNext Ligation Enhancer (red) NEBNext Ligation Enhancer |
1 µl |
 (red) NEBNext Ultra II Ligation Master Mix (red) NEBNext Ultra II Ligation Master Mix |
30 µl |
| Total Volume |
93.5 µl |
The Ligation Master Mix and Ligation Enhancer can be mixed ahead of time and is stable for at least 8 hours @ 4°C. We do not recommend premixing the Ligation Master Mix, Ligation Enhancer and adaptor prior to use in the Adaptor Ligation Step.
1.7.3. Set a 100 μl or 200 μl pipette to 80 μl and then pipette the entire volume up and down at least 10 times to mix thoroughly. Perform a quick spin to collect all liquid from the sides of the tube.
| Caution: The NEBNext Ultra II Ligation Master Mix is very viscous. Care should be
taken to ensure adequate mixing of the ligation reaction, as incomplete
mixing will result in reduced ligation efficiency. The presence of a
small amount of bubbles will not interfere with performance. |
1.7.4. Incubate 15 minutes at 20°C in a thermocycler.
1.7.5 Add 3 μl
 (red) USER Enzyme to the ligation mixture from Step 1.7.4, resulting in total volume of 96.5 μl.
(red) USER Enzyme to the ligation mixture from Step 1.7.4, resulting in total volume of 96.5 μl.1.7.6 Mix well and incubate at 37°C for 15 minutes with the heated lid set to ≥ 45°C.
1.7.7 Proceed immediately to Purification of the Ligation Reaction.
1.8. Purification of the Ligation Reaction Using SPRIselect Beads or NEBNext Sample Purification Beads
| If you are selecting for larger insert size libraries (> 200 nt) follow the size selection recommendations in Appendix A, Section 6. |
1.8.1. Add 87 μl (0.9X) resuspended SPRIselect Beads or NEBNext Sample Purification Beads and mix well on a vortex mixer or by pipetting up and down at least 10 times.
1.8.2. Incubate for 5 minutes at room temperature.
1.8.3. Quickly spin the tube in a microcentrifuge and place the tube on an appropriate magnetic rack to separate beads from the supernatant. After the solution is clear (~ 5 minutes), discard the supernatant that contain unwanted fragments (Caution: do not discard the beads).
1.8.4. Add 200 μl of freshly prepared 80% ethanol to the tube while in the magnetic rack. Incubate at room temperature for 30 seconds, and then carefully remove and discard the supernatant.
1.8.5. Repeat Step 1.8.4 once for a total of 2 washing steps.
1.8.6. Briefly spin the tube, and put the tube back in the magnetic rack.
1.8.7. Completely remove the residual ethanol, and air dry beads until the beads are dry for 5 minutes while the tube is on the magnetic rack with the lid open.
Caution: Do not over-dry the beads. This may result in lower recovery of DNA target. Elute the samples when the beads are still dark brown and glossy looking, but when all visible liquid has evaporated. When the beads turn lighter brown and start to crack they are too dry.
1.8.8. Remove the tube from the magnet. Elute DNA target from the beads by adding 17 μl 0.1X TE (provided) to the beads. Mix well on a vortex mixer or by pipetting up and down. Quickly spin the tube and incubate for 2 minutes at room temperature. Put the tube in the magnetic rack until the solution is clear.
1.8.9. Without disturbing the bead pellet, transfer 15 μl of the supernatant to a clean PCR tube and proceed to PCR enrichment.
| If you need to stop at this point in the protocol, samples can be stored at –20°C. |
1.9. PCR Enrichment of Adaptor Ligated DNA
 Check and verify that the concentration of your oligos is 10 μM on the label.
Check and verify that the concentration of your oligos is 10 μM on the label.
 Use Option A for any NEBNext oligos kit where index primers are supplied in tubes. These kits have the forward and reverse primers supplied in separate tubes.
Use Option A for any NEBNext oligos kit where index primers are supplied in tubes. These kits have the forward and reverse primers supplied in separate tubes.
Use Option B for any NEBNext oligos kit where index primers are supplied in a 96-well plate format. These kits have the forward and reverse (i7 and i5) primers combined.
1.9.1. Set up the PCR reaction as described below based on the type of oligos (PCR primers) used.
1.9.1A. Forward and Reverse Primers Separate
| COMPONENT |
VOLUME |
|---|---|
| Adaptor Ligated DNA (Step 1.8.9) | 15 µl |
 (blue) NEBNext Ultra II Q5 Master Mix (blue) NEBNext Ultra II Q5 Master Mix |
25 µl |
| Index (X) Primer/i7 Primer*, ** | 5 µl |
| Universal PCR Primer/i5 Primer*, ** |
5 µl |
| Total Volume |
50 µl |
* NEBNext Oligos must be purchased separately from the library prep kit. Refer to the corresponding NEBNext Oligo kit manual for determining valid barcode combinations.
** Use only one i7 primer/ index primer per sample. Use only one i5 primer (or the universal primer for single index kits) per sample
1.9.1B. Forward and Reverse Primers Combined
| COMPONENT |
VOLUME |
|---|---|
| Adaptor Ligated DNA (Step 1.8.9) | 15 µl |
 (blue) NEBNext Ultra II Q5 Master Mix (blue) NEBNext Ultra II Q5 Master Mix |
25 µl |
| Index Primer Mix* | 10 µl |
| Total Volume |
50 µl |
* NEBNext Oligos must be purchased separately from the library prep kit. Refer to the corresponding NEBNext Oligo kit manual for determining valid barcode combinations.
1.9.2. Mix well by gently pipetting up and down 10 times. Quickly spin the tube in a microcentrifuge.
1.9.3. Place the tube on a thermocycler with the heated lid set to 105°C and perform PCR amplification using the following PCR cycling conditions (refer to Table 1.9.3A and Table 1.9.3B):
Table 1.9.3A:
| CYCLE STEP |
TEMP | TIME |
CYCLES |
|---|---|---|---|
| Initial Denaturation | 98°C |
30 seconds | 1 |
| Denaturation Annealing/Extension |
98°C 65°C |
10 seconds 75 seconds |
7–15*, ** |
| Final Extension | 65°C |
5 minutes | 1 |
| Hold | 4°C |
∞ |
* The number of PCR cycles should be adjusted based on RNA input (Table 1.9.3B).
** It is important to limit the number of PCR cycles to avoid overamplification.
If overamplification occurs, a second peak ~ 1,000 bp will appear on the Bioanalyzer trace. (See Figure 7.2 in Appendix A).
Table 1.9.3B:
| TOTAL RNA INPUT |
RECOMMENDED PCR CYCLES |
|---|---|
| 1,000 ng | 7–8 |
| 100 ng |
11–12 |
| 10 ng | 14–15 |
Note: PCR cycles are recommended based on high quality Universal Human Reference Total RNA. It may require optimization based on the sample quality to prevent PCR over-amplification.
1.10. Purification of the PCR Reaction using SPRIselect Beads or NEBNext Sample Purification Beads
1.10.1. Vortex SPRIselect Beads or NEBNext Sample Purification Beads to resuspend.
1.10.2. Add 45 μl (0.9X) of resuspended beads to the PCR reaction (~ 50 μl). Mix well on a vortex mixer or by pipetting up and down at least 10 times.
1.10.3. Incubate for 5 minutes at room temperature.
1.10.4. Quickly spin the tube in a microcentrifuge and place the tube on an appropriate magnetic rack to separate beads from the supernatant. After the solution is clear (about 5 minutes), carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets.
1.10.5. Add 200 μl of freshly prepared 80% ethanol to the tube while in the magnetic rack. Incubate at room temperature for 30 seconds, and then carefully remove and discard the supernatant.
1.10.6. Repeat Step 1.10.5 once for a total of 2 washing steps.
1.10.7. Air dry the beads for 5 minutes while the tube is on the magnetic rack with the lid open.
Caution: Do not over-dry the beads. This may result in lower recovery of the DNA target. Elute the samples when the beads are still dark brown and glossy looking, but when all visible liquid has evaporated. When the beads turn lighter brown and start to crack they are too dry.
1.10.8. Remove the tube from the magnetic rack. Elute the DNA target from the beads by adding 23 μl 0.1X TE (provided) to the beads. Mix well on a vortex mixer or by pipetting up and down ten times. Quickly spin the tube in a microcentrifuge and incubate for 2 minutes at room temperature. Place the tube in the magnetic rack until the solution is clear.
1.10.9. Transfer 20 μl of the supernatant to a clean PCR tube, and store at –20°C.
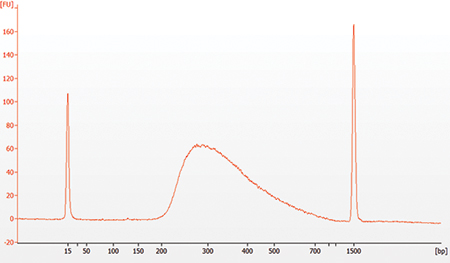
1.11 Assess Library Quality on a Agilent Bioanalyzer DNA Chip
1.11.1. Run 1 μl library on a DNA 1000 chip. If the library yield is too low to quantify on this chip, please run the samples on a DNA High Sensitivity chip. A dilution may be necessary for running on a Bioanalyzer High Sensitivity DNA chip.
1.11.3. Check that the electropherogram shows a narrow distribution with a peak size approximately 300 bp.
Note: If a peak at ~ 80 bp (primers) or 128 bp (adaptor-dimer) is visible in the Bioanalyzer traces, bring up the sample volume (from Step 1.10.9) to 50 μl with 0.1X TE buffer and repeat the SPRIselect Bead or NEBNext Sample Purification Bead Cleanup Step (Section 1.10).