Ph.D.™-C7C Phage Display Peptide Library Kit
This product was discontinued on December 15, 2022 and is replaced with Ph.D.-C7C Phage Display Peptide Library Kit v2 (NEB #E8212).
- Replaced by PhD-C7C Phage Display Peptide Library Kit v2 on December 15, 2022
- Catalog # E8120 was discontinued on December 15, 2022
The Ph.D.™-C7C Phage Display Peptide Library Kit contains a tube of the Ph.D.-C7C Phage Library, protein reagents for a control reaction and sequencing primer stocks. The Ph.D.-C7C library is a combinatorial library of random peptides with a disulfide constrained loop, fused to a minor coat protein (pIII) of M13 phage. The displayed peptide, in the form AC-X7-CGGGS, is expressed at the N-terminus of pIII. The library consists of approximately 1 x 109 electroporated (i.e., unique) sequences.
- Ready to use complex phage peptide library
- Structurally constrained peptide library
- Applications and methodologies are numerous and supported by over 20 years of literature citations
- Does not require helper phage for amplification
- Inherent link between phenotype and genotype allows screening of billions of clones in a single microtiter well or Eppendorf tube

-
Product Information
The Ph.D.-C7C Phage Display Peptide Library is based on a combinatorial library of random peptides fused to a minor coat protein (pIII) of M13 phage (1–6). The displayed peptide is expressed at the N-terminus of pIII, i.e., the first residue of the mature protein is the first randomized position. The peptide is followed by a short spacer (Gly-Gly-Gly-Ser) and then the wild-type pIII sequence. The library consists of approximately 109 electroporated sequences amplified once to yield approximately 100 copies of each sequence in 10 µl of the supplied phage.
Please click the link for Applications of the Ph.D. Phage Display Peptide Libraries.
Complexity: 1.2 x 109 transformants.
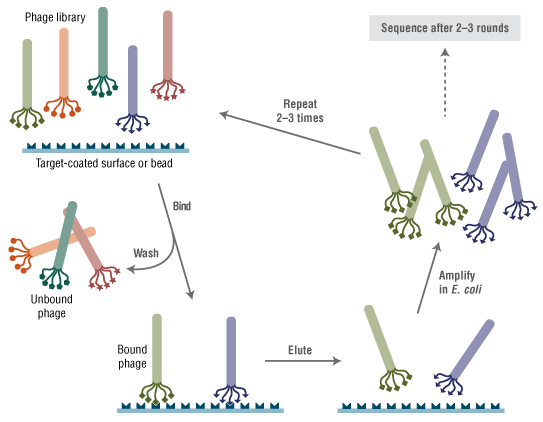
Panning with a pentavalent peptide library displayed on plll. - This product is related to the following categories:
- Discontinued (<3 years)
-
Protocols, Manuals & Usage
-
FAQs & Troubleshooting
-
Citations & Technical Literature
-
Quality, Safety & Legal
Other Products You May Be Interested In
Ineligible item added to cart
Based on your Freezer Program type, you are trying to add a product to your cart that is either not allowed or not allowed with the existing contents of your cart. Please review and update your order accordingly If you have any questions, please contact Customer Service at freezers@neb.com or 1-800-632-5227 x 8.


