Omicron Update
The Luna SARS-CoV-2 RT-qPCR assay (NEB #E3019) is an RUO multiplex assay based on the US CDC-designed SARS-CoV-2 N1 and N2 genes. The new COVID-19 variant, Omicron, contains one point mutation in a region of the N-gene that is targeted by the N1 probe (C28311U). Direct testing of purified RNA representing both the wild-type (Wuhan) and mutant SARS-CoV-2 (Omicron) N1 templates resulted in substantially equivalent results and 100% detection of both samples at 10 copies per reaction (27/27 replicates tested).
Additionally, a subpopulation of the Omicron BA.5 variant contains an additional point mutation at the 3′ end of the N1 probe (A28330G). Testing of purified RNA representing both the wild-type (Wuhan) and mutant BA.5 template (C28311U, A28330G) also resulted in substantially equivalent results.
Other Variants
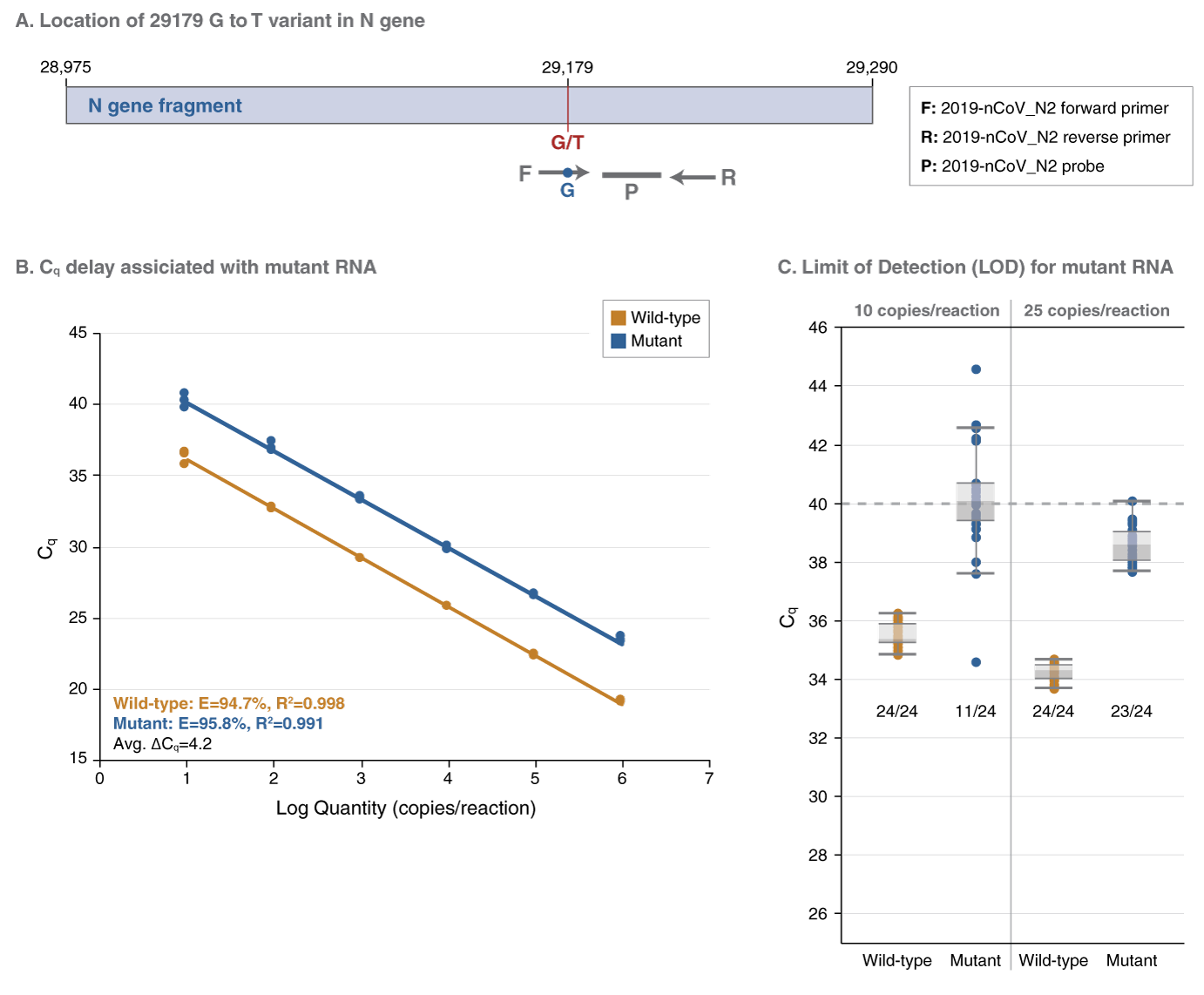
NEB has developed a web tool (Primer Monitor) to continually monitor registered primer sets for overlapping variants in sequences from GISAID. Using this tool, we identified a prominent variant from some countries with a SNP close to the 3’ end of the 2019-nCoV-2_N2 forward primer included in the Luna kit, which detects the CDC SARS-CoV-2 N1, N2 targets and human RNase P gene with modifications (click for details) (A). To evaluate the impact of this SNP on the N2 target detection, we prepared two N gene RNA fragments containing the wild type and mutant N2 targets, respectively, by in vitro transcription and quantitated them using the SARS-CoV-2 RNA Control 2 from Twist Bioscience. Using the primer/probe set included in the Luna kit, we observed an average 4.2 Cq delay for the mutant N2 target (B). The limit of detection (LOD) for the mutant N2 target was 25 copies/reaction. Though all the reactions containing 10 copies of the mutant RNA generated amplification signal, only 11 of 24 had a Cq ≤ 40, the cut-off for detection (C).