NEBioCalculator® - Using the ds: mass -> ends module to plan a phosphorylation reaction using T4 Polynucleotide Kinase (T4 PNK)
Script
NEBioCalculator is an easy-to use tool that helps with various biomass calculations. The double-stranded DNA, mass-to-ends module is useful when setting up a phosphorylation reaction using T4 Polynucleotide Kinase, also known as T4 PNK, because it converts linear double-stranded DNA mass to moles of DNA ends.
T4 PNK will phosphorylate up to 300 picomoles of 5 DNA ends in a single reaction. Nucleotide concentration is often provided in mass concentration, such as nanograms per microliter, rather than a quantity of total ends in solution. NEBioCalculator can help to make this conversion by entering DNA length in base pairs or kilobases, and the mass of DNA in micrograms, nanograms, etc., in the reaction. To do this, enter the DNA length and the mass of the DNA in the reaction. Please make sure to select the correct units from the pull-down.
In the following example, we calculate the number of ends for a 200 base pair double-stranded DNA fragment that we need to phosphorylate. Our example uses 10 microliters of a 50 nanogram per microliter DNA solution, or 500 nanograms total. NEBioCalculator immediately calculates the number of moles of DNA ends that we have in the reaction to be 8.089 picomoles.
Since one microliter of T4 PNK can phosphorylate up to 300 picomoles of five-prime end termini, we recommend using one microliter of enzyme in a reaction volume up to 50 microliters.
Please note, that this calculation is performed using the average mass of the standard ATCG nucleotides. NEBioCalculator can use your DNA sequence to calculate the expected mass of your specific sequence. Just paste your sequence into the optional sequence box at the top. The DNA length will automatically update to reflect what you have entered.
Find more of our helpful NEBioCalculator modules at nebiocalculator.neb.com.
Related Videos
-
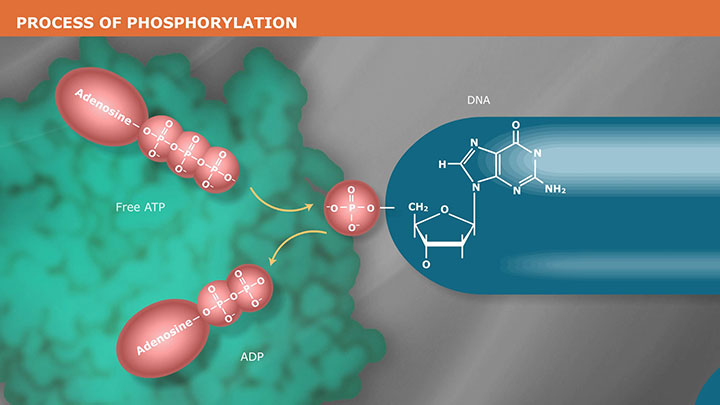
The Mechanism of DNA Phosphorylation -

Quick Tips - Do I need a 5' phosphate for ligation? -
NEBioCalculator® - Using the Ligation module

