
Overcoming challenges in FFPE DNA library prep
Posted on Wednesday, October 25, 2023
By
Topic: Tips for the lab, What is Trending in Science
Formalin-fixed paraffin-embedded (FFPE) samples are a critical resource for clinicians and scientists, with valuable sequencing insights that has been limited by challenges to library prep. FFPE samples are specimens that have been preserved for the long haul. The formalin-fixation and paraffin-embedding process allows hospitals to store these samples at room temperature, making them easily accessible for analysis by pathologists. More recently, there has been a growing interest in extracting DNA from FFPE samples for genetic analysis. But, attempting to prepare DNA libraries for sequencing from FFPE samples can be particularly challenging due to the effects of formalin fixation and paraffin embedding on the quality of DNA.
What are FFPE samples?
Formalin fixation is a commonly used method for preserving tissue specimens, which involves immersing them in a solution containing formaldehyde. This stabilizes the tissue, preventing decay and preserving architecture for examination under a microscope. However, formalin fixation also leads to chemical modifications of DNA, and crosslinks between nucleic acids and proteins, making it more difficult to extract intact DNA from the tissue.
The paraffin embedding process used in conjunction with formalin fixation enables sectioning of samples yet further degrades nucleic acids. This involves infiltrating the tissue with molten paraffin wax, solidifying and forming a protective barrier around the tissue. While this method provides excellent preservation of tissue architecture, it also causes physical damage to DNA due to heat and dehydration.
As a result of these processes, FFPE samples often provide highly degraded DNA with low yields, making it challenging for researchers to obtain enough high-quality DNA for genetic analysis, such as whole genome sequencing and mutation analysis. Advancements in developing specialized techniques and kits for DNA repair and fragmentation have paved the way for researchers to unlock valuable genetic insights, even from samples deemed too compromised to yield meaningful results.
Challenges with FFPE samples: Low input, non-uniform ends and DNA damage
When using FFPE sample-extracted DNA, researchers face challenges such as low input amounts, non-uniform ends and damaged bases (1). These DNA outcomes hinder library preparation and downstream analysis.
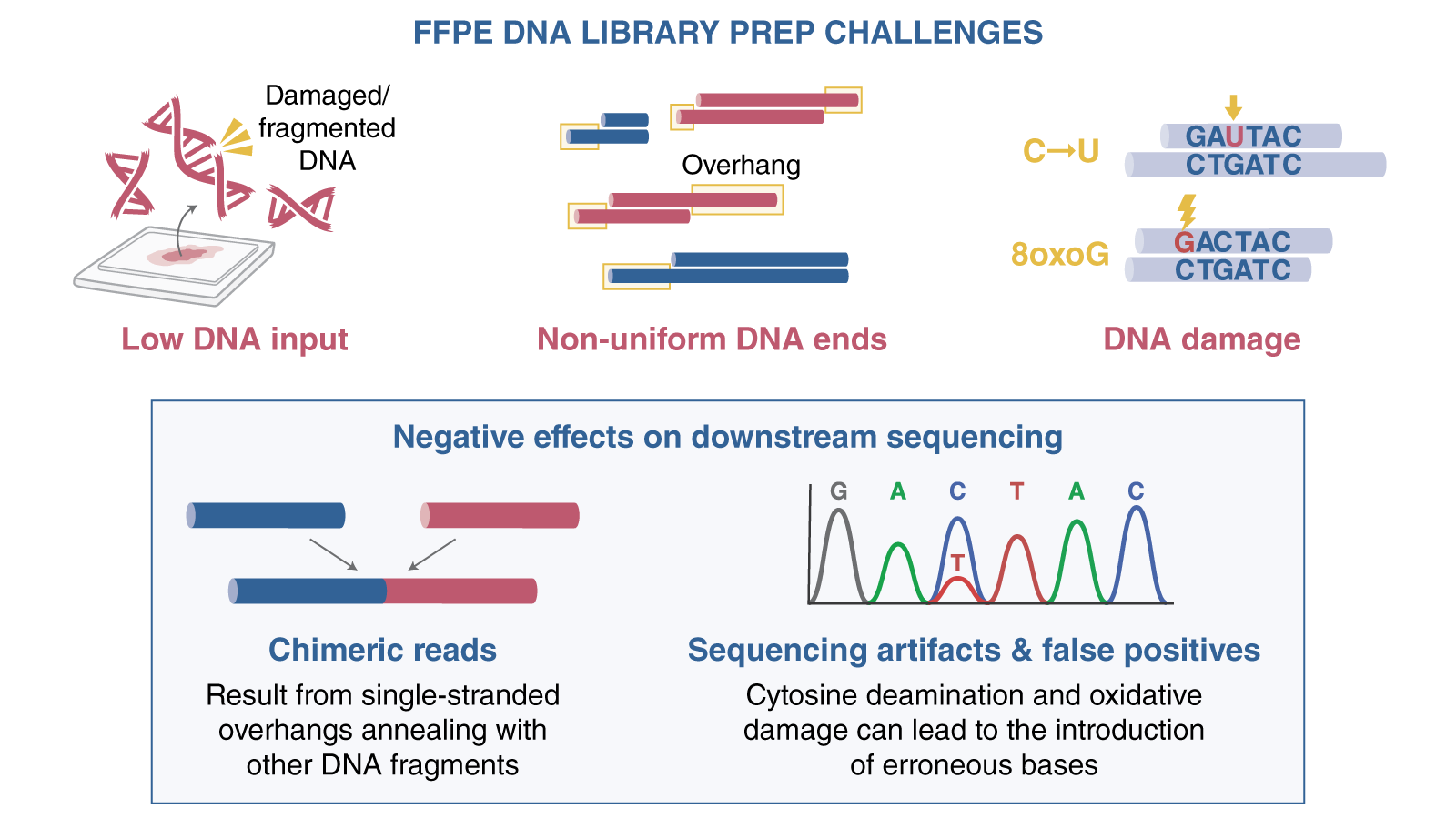
Figure 1: Some of the challenges faced by researchers when using DNA extracted from FFPE samples are low input, non-uniform ends and DNA damage.
- Low input amounts: results in part because the available tissue sample is minute and the DNA extracted is fragmented and damaged. Consequently, only a limited quantity of usable DNA is available for library prep workflows.
- Non-uniform ends: can result from nicking, leading to challenges during library preparation.
- Damaged DNA: FFPE samples exhibit distinct types of damage, including cytosine deamination (C to T mutation) and oxidative damage (e.g., 8-oxo G leading to G to T mutation). Other types of damage include nicks, gaps, and abasic sites, which can hinder library yield due to polymerase blockage.
These challenging DNA samples can cause the following problems for downstream sequencing: - Chimeric reads: Single-stranded overhangs can anneal with other DNA fragments, leading to chimeric reads during sequencing.
- Sequencing artifacts: Damage to bases caused by cytosine deamination and oxidation in single-stranded regions can cause problems during end repair by introducing erroneous bases, resulting in sequencing artifacts; when researchers search for mutations in patient samples, these artifacts can lead to false-positive results.
Concerns with Enzymatic Fragmentation
Enzymatic and mechanical fragmentation are vital techniques for processing DNA from formalin-fixed paraffin-embedded (FFPE) samples. Enzymatic methods involve using nucleases to cleave DNA, while mechanical techniques rely on physical forces like sonication. Historically, enzymatic fragmentation methods faced concerns, such as:
- Bias: Some enzymes exhibit bias in DNA fragmentation due to sequence-specific cleavage, leaving certain genome regions underrepresented in sequencing data.
- Inconsistent sizing: DNA fragments produced by various enzyme mixes pose challenges in controlling the DNA fragment size.
- Overfragmentation: A common concern with FFPE samples is the fear of complete digestion to the point of unusable fragments, but research at NEB indicates that prolonged fragmentation time doesn't significantly alter the size of small, pre-fragmented DNA in FFPE samples.
NEBNext UltraShear™ FFPE DNA Library Prep Kit utilizes many unique enzyme qualities to mitigate FFPE library prep issues
Our NEBNext portfolio of products focuses on streamlined workflows and kits for next generation sequencing of many sample types. Our standard library prep kits work very well with DNA extracted from cell lines, frozen tissues and even cell-free DNA, but FFPE is a different beast.
The recently released NEBNext UltraShear FFPE DNA Library Prep Kit (NEB #E6655) dramatically improves the process of library preparation from FFPE tissue samples by employing a time-dependent DNA fragmentation method primarily based on a specialized enzyme mix. The repair and fragmentation workflow improves sequence complexity and coverage uniformity from DNA derived from FFPE samples, offering a more comprehensive representation of the genomic content of the sample.
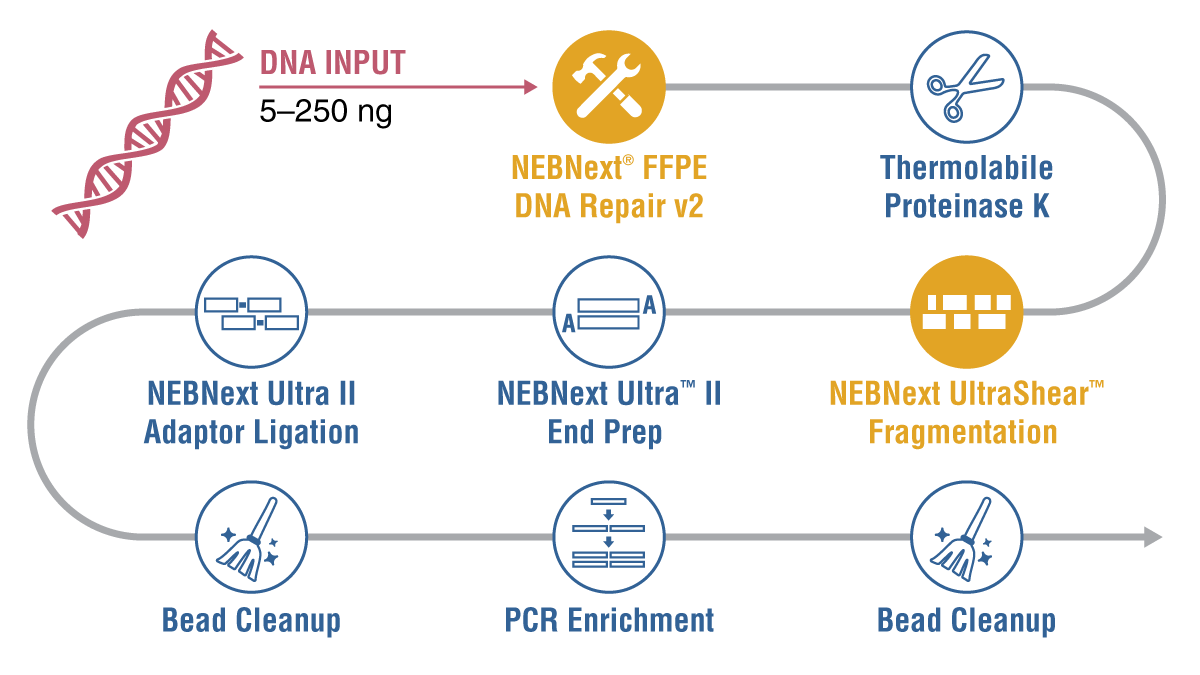
Figure 2: The NEBNext UltraShear FFPE DNA Library Prep Kit has a streamlined workflow with minimal hands-on time across a range of input amounts.
The workflow begins with the NEBNext FFPE DNA repair V2 mix, which selectively targets damaged DNA bases. When single-stranded damage is detected, the damaged portions are excised, while double-strand damage undergoes base excision repair mechanisms. This significantly enhances data accuracy by removing artifacts resulting from damaged sites, and it boosts library conversion rates by rectifying issues such as nicks, gaps, and overhangs, leading to superior library quality.
The repair step also:
- Prevents over-fragmentation: Repairing nicks and gaps before fragmentation helps maintain the DNA's original fragment size, preventing further size reduction during fragmentation.
- Retains intact DNA: Some single-stranded overhangs don't have any damage. Repairing by filling those overhangs in retains intact DNA for incorporation into the library; otherwise, sequencing coverage would be diminished.
- Preserves mutations: Because the repair step specifically targets damaged bases (such as cytosine deamination and oxidative damage, as mentioned above), true mutations are left intact. True mutations appear on both DNA strands and damaged bases appear only on one strand and can be safely removed. An essential consideration in developing repair & fragment workflows is that polymerase activity should occur AFTER damaged bases have been removed; otherwise, in FFPE samples, they can result in much higher levels of the false positives mentioned above. Many researchers working with FFPE samples need to enrich particular sites of interest for mutation analysis due to poor coverage uniformity. Without target enrichment, whole genome sequencing becomes too expensive. The NEBNext Ultra Shear FFPE DNA Library Prep Kit gives the best yields and highest on-target reads even from extremely low-quality samples.
A Sample-Quality-Agnostic Workflow for FFPE Samples
The NEBNext UltraShear FFPE DNA Library Prep Kit provides a largely sample quality-agnostic workflow with a broad input range. This allows customers to apply a single protocol condition across a whole plate of samples regardless of the quality or input amount. This is helpful in high throughput settings, such as clinical labs. A significant challenge for clinical labs is handling different sample types (high-quality vs. FFPE) without requiring distinct workflows. Often, clinical labs don't have the ability (such as time and sample quantity) to characterize or tweak conditions based on different sample quality. Enzymatic solutions are also very automation friendly. Enzymatic fragmentation offers the advantage of being less dependent on sample quality and resistant to over-fragmentation, making it a viable choice for both high- and low-quality samples.
NEBNext UltraShear™ FFPE DNA Library Prep has significant workflow and reagent advancements for working with FFPE samples in molecular labs that address the challenges associated with library preparation. Researchers now have a robust method to improve library quality and extract meaningful genetic insights from FFPE samples, irrespective of their quality or quantity.
Learn more about NEBNext solutions for working with FFPE DNA.
1. Haile, S. et al. (2019) Nucl. Acids Res. PMID: 30418619
NEB will not rent, sell or otherwise transfer your data to a third party for monetary consideration. See our Privacy Policy for details. View our Community Guidelines.
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. See www.neb.com/trademarks. The use of these products may require you to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
Don’t miss out on our latest NEBinspired blog releases!
- Sign up to receive our e-newsletter
- Download your favorite feed reader and subscribe to our RSS feed
Be a part of NEBinspired! Submit your idea to have it featured in our blog.


