Site Directed Mutagenesis Site-directed mutagenesis — experimental considerations
Site-directed mutagenesis — experimental considerations
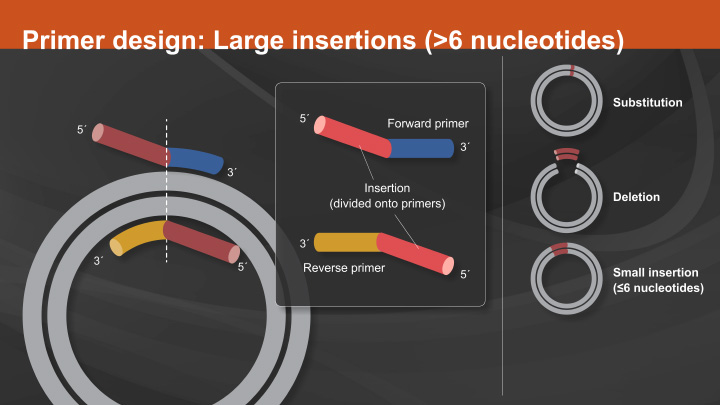
Return to Site Directed MutagenesisPrimer Design:
The most critical component to the success of a SDM experiment is proper primer design. As mentioned above, the first consideration when designing primers is the location of the two primers in relation to each other. Primers designed back-to-back have the benefit of exponential amplification. However, polymerase errors during PCR will also be propagated exponentially, therefore only the highest fidelity enzymes should be used.
Another important aspect to consider when designing primers is the melting temperature. The forward and reverse primers should be designed with similar melting temperatures so that each primer anneals with comparable efficiency. Melting temperature calculations can be challenging for SDM as most online tools do not account for alterations in annealing temperatures due to mismatched nucleotides.
In contrast, NEBaseChanger, a free online primer design tool, has been developed not only to aid in the design of primers for the Q5 Site-Directed Mutagenesis Kit, but also to provide an annealing temperature for Q5 that accounts for mismatched nucleotides.
Primer Modification:
For some workflows, primers must be synthesized with a 5-prime phosphate to enable a downstream intramolecular ligation reaction (this is not required for the Q5 Site-Directed Mutagenesis Kit.
Primer Purity:
An additional consideration is the purity of the primers. Incomplete primer synthesis can lead to errors at the mutagenesis site. As synthesis errors occur more often with larger primers, it is recommended to have primers > 40-50 nucleotides long, PAGE purified.
Template Removal:
For most SDM experiments, the non-mutated PCR template is removed from the pool of PCR products using the restriction endonuclease DpnI. DpnI selectively digests methylated DNA (i.e. plasmids propagated and isolated from E. coli). Because PCR products are generated in vitro, they do not contain methyl groups and are resistant to DpnI activity.
Ligation:
Back-to-back primer design results in a linear double-stranded PCR product. Prior to transformation, this product is circularized using an intramolecular ligation reaction. Given the proper buffer conditions, this phosphorylation and ligation reaction can be accomplished in as few as five minutes at room temperature.
Transformation:
Products of SDM can be transformed into most chemically-competent E. coli strains suitable for cloning. Transformation efficiency will vary depending on the size of the plasmid as well as the quality and efficiency of the competent cells. Electroporation is possible if the salt content of the plasmid solution is removed via dialysis or buffer exchange. For protocols that include the transformation of unligated plasmids, transformation efficiencies are decreased.
Product evaluation:
In order to ensure the success of the SDM reaction, the final plasmid should be isolated from a single E. coli clone and the site of mutation should be sequenced in both directions. As long as the fidelity of the polymerase used is sufficient, it is usually not necessary to sequence the entire plasmid.
Videos
-

NEBaseChanger® Primer Design Tool Tutorial
-
Overview of the Q5® Site-Directed Mutagenesis Kit
Learn how to create substitutions, deletions or insertions in 3 easy steps with the Q5 Site-Directed Mutagenesis Kit.
-
Troubleshooting tips for Q5® Site-Directed Mutagenesis Kit
Tips for commonly encountered challenges in site-directed mutagenesis.

