NEBaseChanger® Primer Design Tool Tutorial

Script
Narrator:
The NEBaseChanger tool allows users to design primers to be used with the NEB Q5 Site-Directed Mutagenesis Kit or with NEBuilder HiFi DNA Assembly for multi-site mutagenesis applications only. We will first demonstrate how to use NEBaseChanger to design primers for single site-directed mutagenesis for use with the Q5 Site-Directed Mutagenesis Kit. These can be amino acid changes or other sequence insertions, deletions, or substitutions. To get started, enter or select your sequence.
FASTA or raw text files can be uploaded, or you can choose a common vector sequence from the list in the sample vectors drop-down menu. Once the sequence is populated, you can select which type of mutation is to be programmed. If changing an amino acid codon or programming an insertion, deletion, or substitution, a primer set ID may be entered if desired. We will now look at each mutation programming option in turn. To program an amino acid change, the open reading frame start position must be set.
Once this is set, the mutation is entered in the enter mutations field in the following format: old amino acid, amino acid number, new amino acid. The system will check to ensure that the old amino acid matches the codon position entered. In the event that it is incorrect, a warning will display. To enter a stop codon, use an asterisk in place of a new amino acid. Choose a codon selection strategy, either maximum codon usage or maximum parsimony. For the former, the codon with the highest frequency usage for each amino acid from the E. coli K-12 is applied.
For maximum parsimony, the sequence distance is calculated between the original codon and all mutant codons that encode for the desired amino acid and the number of base changes needed to convert the two is minimized. Here we will choose the codon usage for codon selection. Multiple amino acid changes can be programmed at one time by entering them in the same field. A distinct primer set for each of these changes will be generated and displayed as a downloadable table under required primers.
The reading frame is shown above the primer sets and will shift to the appropriate sequence area as you toggle between primer sets. You may choose to confine the location of the mutations within the primer to the five prime tails by clicking the appropriate box. If this option is selected, the mutagenic sequence will always be attached to the five prime end of the primers. This feature is particularly useful for producing primers that allow you to adjust to have the same melting temperature, simplifying PCR for a large batch of mutations at the same site.
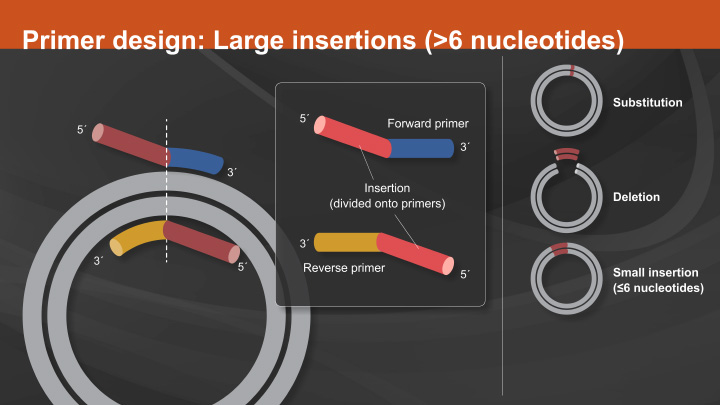
When programming an insertion, deletion, or substitution, the second input format box is selected. If the sequence to be changed or removed is known, you can search for that sequence using the search field. If there are multiple matching sequences found, you can scroll between them to select the correct one that is to be changed. Alternatively, if the start and end positions are known, you can enter them manually. If programming a substitution, the new sequence to replace that which is highlighted will be entered into the replacement sequence box.
You may substitute with a sequence that is longer than the original sequence. For example, when replacing a codon with multiple codons. The primers reflecting those substitutions will be displayed below. If programming an insertion, the start and end positions should be consecutive. The replacement sequence will be inserted between them. For convenience, if inserting a tag, you can select one from the common peptide tags. Or if inserting an amino acid or stop codon, you can select one from the drop-down menu.
You can also type in the sequence to be inserted. Again, primers will be displayed below. When programming a deletion, enter the base positions flanking the sequence to be deleted and simply leave the replacement sequence field blank. Resulting primers will be shown below. Finally, if multiple different types of changes are to be made on an input sequence, batch mode can be used. Batch input should be formatted as either amino acid changes or insertion, deletion operations.
All batch input operates on the currently loaded sequence. For amino acid mutations, the start of the open reading frame must be indicated by entering ORF and the position of the start. The next line can indicate the amino acid changes in the format old amino acid, amino acid number, new amino acid. Each mutation must be on a separate line and all mutations are referenced relative to the open reading frame start. A new open reading frame can be specified by inserting another line, ORF: new start position.
All subsequent mutations will be relative to this new open reading frame start. Insertions, deletions, and substitutions are formatted as an identifier, the start position of the change, the end position of the change, and the replacement sequence. When programming a substitution, the sequence between bases 234 and 239 will be replaced with ATCG. For deletions, the replacement sequence part of the line is left blank. Here, the sequence between bases 234 and 239 will be deleted.
For insertions, the start and end positions should be consecutive. Here, ATCG will be inserted between bases 234 and 235. Once all desired changes are listed, select build primers to display the generated primer sets. Whether you are programming amino acid changes, insertions, deletions, or substitutions, or using batch mode, you can download all primer sets by selecting download all primers. The resulting plasmid sequence incorporating all the changes can be downloaded by using the download selected sequence button.
Additionally, there is a summary table that can be downloaded that shows the primers, their lengths and characteristics, as well as the changes being made. Now let's walk through performing multi-site directed mutagenesis using NEBuilder HiFi DNA Assembly. NEBaseChanger enables the creation of primers to generate multiple point mutations in your DNA simultaneously in a single reaction using the assembly kit rather than the Q5 mutagenesis kit. Start by inputting your sequence.
Here, we will input a circular vector sequence containing the gene of interest, which we'll be making point mutations in. Next, select amino acid point mutation and specify the start position of the open reading frame. The mutagenesis sites will be determined relative to this start. In this plasmid, the gene of interest starts at nucleic acid position 266. Next, enter one or more of the desired mutations in the form of old amino acid, amino acid number, new amino acid. Here, we will be making mutations V245F, E462S, and N726F.
Select a codon selection strategy, either maximum codon usage or maximum parsimony. Now, check the box to generate primers for NEBuilder HiFi and select multi-site reaction. When multi-site is selected, primer pairs will be generated to produce fragments by PCR that can be assembled in the NEBuilder HiFi reaction, modifying multiple sites in a single reaction and creating a single defined multi-mutant. Finally, click the build primers button to obtain the table of primers.
Here, we have three primer pairs that will be used to create three PCR generated fragments containing the desired mutations. The first fragment is 674 base pairs long, the second, 818, and the third, 5,801. This fragment is longer because it spans the length of the vector sequence. You're now ready to assemble your PCR fragments in an NEBuilder HiFi DNA Assembly Reaction. Note: if single site is selected, primer pairs will be generated for each site independently and grouped according to each individual mutation.
Users interested in generating combinatorial libraries can mix the forward and reverse primers of different mutations to create the custom fragments needed for NEBuilder HiFi Assembly Reactions. If you have any questions relating to the use of the new NEBaseChanger tool, please don't hesitate to reach out to technical support at info@NEB.com.
Related Videos
-
Overview of the Q5® Site-Directed Mutagenesis Kit -
Troubleshooting tips for Q5® Site-Directed Mutagenesis Kit -

Quick Tips - Can I make multiple mutations with the Q5® site-directed mutagenesis kit?

