Overview of NEBridge® Tools for Golden Gate Assembly

Script
In this video, we will introduce you to the multiple web tools that New England Biolabs offers to assist you with designing high-fidelity, high-efficiency Golden Gate Assemblies using NEB Type IIS restriction enzymes. We will provide an overview of our available web tools, including the NEBridge Golden Gate Assembly Tool and the NEBridge Ligase Fidelity Tools, to help you understand what they do and when to use them.
The NEBridge Golden Gate Assembly Tool (found at goldengate.neb.com) simplifies in silico DNA construct design. We recommend using this tool to visualize your assembly designs and confirm the desired final sequence. The Golden Gate Assembly tool is used to arrange the insert fragments in the correct order, select fusion site overhangs, and also flag any Type IIS recognition sites within your fragment sequences for the enzyme used in your design. These additional sites need to be removed for efficient assembly. This is a process called domestication.
The tool provides primer recommendations to generate your fragment sequences via PCR. It appends the Type IIS binding sites to generate the required overhangs. Overhang sequences are selected to maximize the expected ligation fidelity of the assembly. A detailed video tutorial for the NEBridge Golden Gate Assembly Tool is available to assist you in planning your Golden Gate Assemblies. The NEBridge Golden Gate Assembly Tool also contains the SplitSet-Lite Golden Gate Utility. This feature splits long DNA sequences into smaller fragments at optimal breakpoints for Golden Gate assembly and provides primer recommendations. These fragments may then be used in the main Golden Gate Assembly tool to verify assembly order. SplitSet-Lite is suitable for less than or equal to 20 fragments. For more complex assemblies, the full SplitSet feature is available with the NEBridge Ligase Fidelity tools.
The NEBridge Ligase Fidelity tools can be found at ligasefidelity.neb.com. These tools allow assemblies to be checked for potential assembly fidelity and efficiency issues. They support the design of new assembly standards, expansion of existing assemblies, as well as complex, high fragment assemblies. The suite of tools includes NEBridge Ligase Fidelity Viewer, NEBridge GetSet, and NEBridge SplitSet, and each has its own unique utility. Let’s take a look at each of the tools and how it is used.
The NEBridge Ligase Fidelity Viewer helps users evaluate their existing designs by estimating the fidelity of an overhang set. It can be used to identify ligase fidelity issues with an existing assembly, and it will provide a visualization of overhang ligation preferences. Here we specify our overhang length, ligation conditions, Type IIS enzyme, and the overhangs used in our design. The output provides an estimated ligation fidelity and a matrix visualization. In this example, using the given set of overhangs, Golden Gate Assembly is predicted to yield 95% of correctly ligated products. When a low fidelity pairing is found, the overhangs that will result in mis-ordered fragment assembly due to mismatches are flagged.
The NEBridge SplitSet Tool splits a gene, or other DNA sequence, into multiple fragments at optimized fusion sites, enabling scarless high-fidelity assembly. It permits fusion sites to be limited to specific regions or precise locations. The tool can also assist in selection of fusion sites appropriate for making mutations (including domestication) by incorporation of desired mutations into primers. For example, here we demonstrate splitting a gene into 23 fragments. We prompt SplitSet with our desired Type IIS restriction enzyme, incubation condition, the number of desired fragments, and the DNA sequence to split. We can choose to exclude any overhangs or modify the regions where the sequence will be divided. In this example, the outputs are an optimal overhang set with 99% ligation fidelity, the DNA sequence overhang coordinates, and the sequence fragments. SplitSet can be used with any number of fragments. However, when working with fewer than 20 fragments, we suggest using SplitSet-Lite, which is included as part of the NEBridge Golden Gate Assembly Tool as mentioned previously.
We hope you find this suite of online tools useful for your Golden Gate assemblies.
Please visit neb.com/goldengate for additional resources and tutorials.
Related Videos
-

Golden Gate Assembly Tool Tutorial -
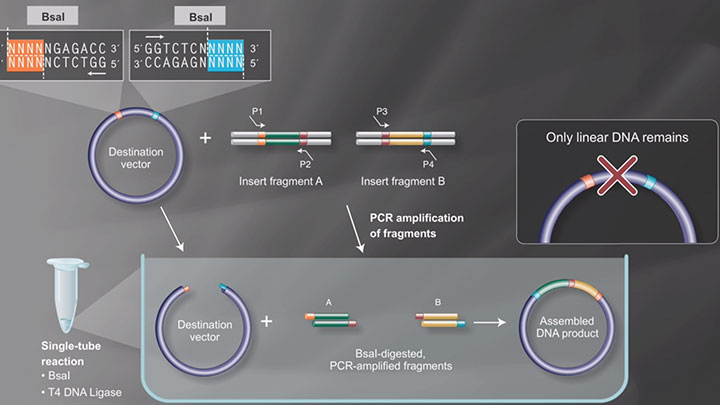
Golden Gate Assembly Workflow -
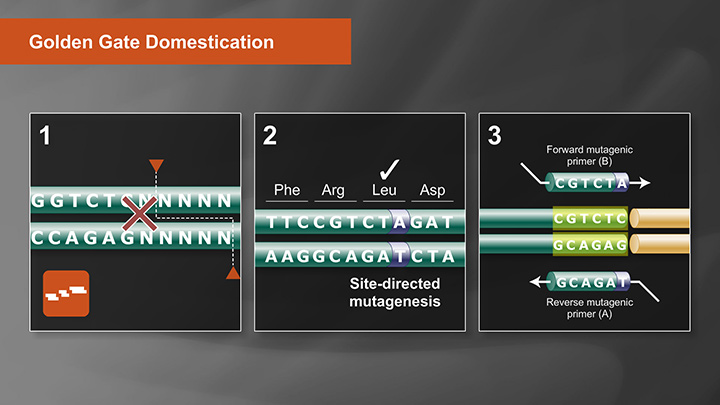
Golden Gate Assembly Domestication Tutorial -

Listen to DAD (Data-optimized Assembly Design) when constructing high-complexity Golden Gate Assembly targets

