NEB® TV Ep. 12 – Applications of DNA Assembly
Script
NEB TV: What's Trending in Science.
Deana Martin, PhD:
Welcome to NEB TV. Today, I'm joined by Tom Evans who is the Scientific Director of our DNA Enzymes Division. Hey, Tom.
Tom Evans, PhD:
Hi, Deana. How are you?
Deana Martin, PhD:
Good. And today, we're talking about applications of DNA assembly. In our Science in 60, Tom will give us an overview about how DNA assembly works. Then, we'll hear from some of our customers about how they use DNA assembly. And lastly, we'll show you how DNA assembly can be used to synthesize sgRNA. You ready?
Tom Evans, PhD:
I'm ready. This will be fun.
Deana Martin, PhD:
Great.
Science in 60
Tom Evans, PhD:
The discovery of enzymes and the development of techniques to accurately cut, move and join distinct DNA molecules revolutionized the life sciences. Traditionally, restriction enzymes and DNA ligases were used to cut and join these DNA molecules in a sequential manner.
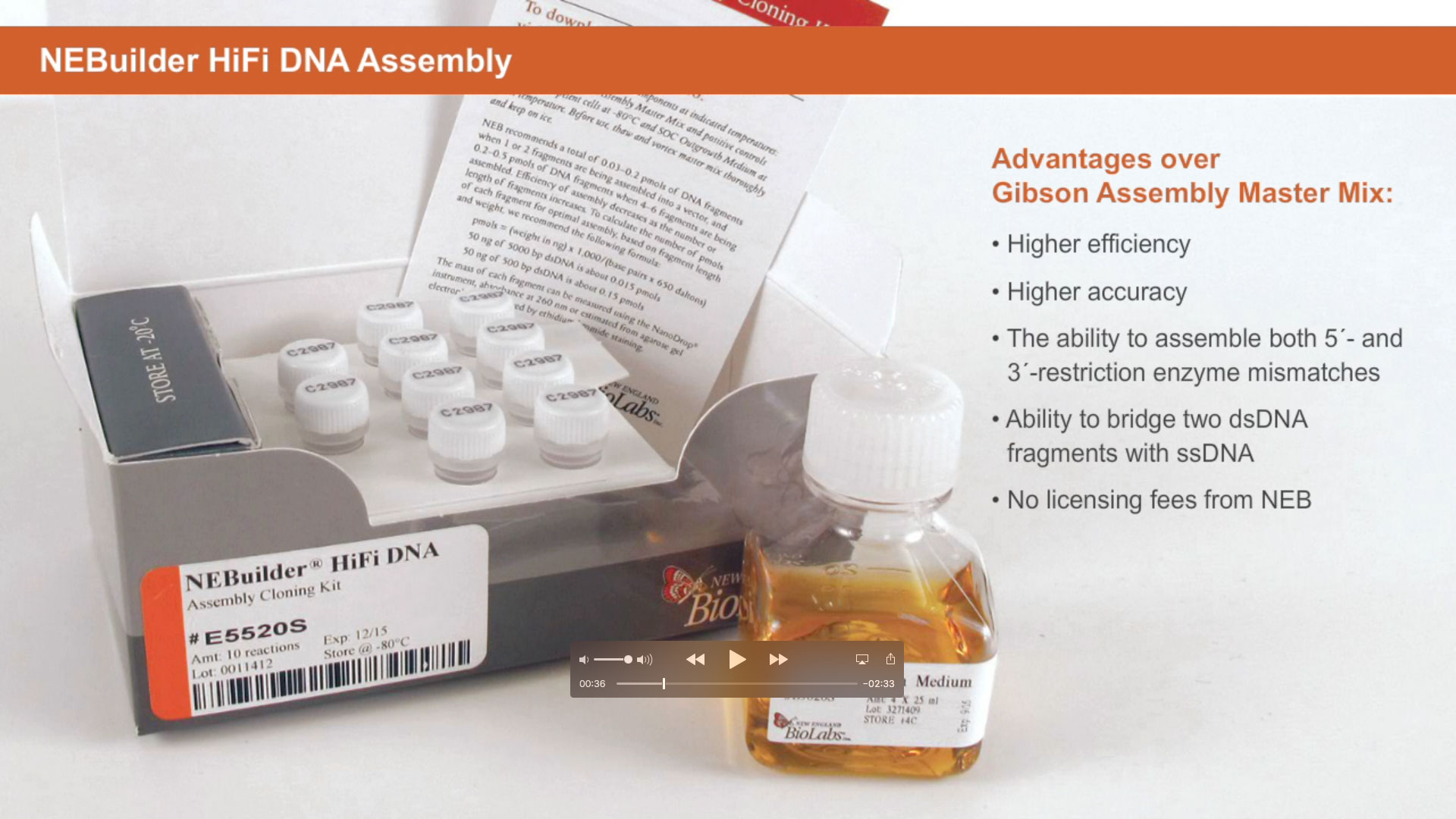
Despite the popularity of traditional techniques, other methods were subsequently developed. DNA assembly, which can be performed using our NEBuilder HiFi products can assemble linear DNA fragments generated by PCR, restriction digests or chemical synthesis.
These fragments need regions of homology in order to give specificity to the reaction. The process starts when a 5' to 3' exonuclease exposes single stranded regions on each of the DNA fragments. The newly created single stranded regions anneal to their complement on a separate DNA fragment. Any remaining gaps are filled in by a DNA polymerase, and the joining is completed when a ligase covalently attaches the newly assembled DNA fragments.
Fast, accurate and simple to use, DNA assembly can be used for a whole variety of applications.
Applications of DNA Assembly
For what types of assemblies are you using NEBuilder HiFi?
Greg Foley, Senior Strain Engineer, ENEVOLV:
So, one thing that I do quite frequently is clone one new guide RNA into the same guide RNA backbone, and it works for that. And the other thing that we do is just make a whole new plasmid so that would take six or seven pieces from many different plasmids and stitch them together into a completely new backbone.
Yao Zhang, PhD, MIT:
Right now, I use NEBuilder actually for all of my clonings. These include complex situations when I have to assemble three or five pieces into one plasmid, or just even one single piece into one backbone.
IIan Wapinski, PhD, Director of Development, ENEVOLV:
I sometimes just put one piece together with a backbone and I just use it with NEBuilder to design the primers because I know it's gonna work.
Why use NEBuilder HiFi DNA Assembly?
Yao Zhang:
Most importantly, it simplifies the cloning procedure dramatically and it's greatly sped up my product and saved me a lot of time.
IIan Wapinski:
It's incredibly reliable. So, because we know it's gonna work, it cuts down on the amount of downstream screening that we have to do afterwards.
Yao Zhang:
In some cases, it's enabled me to create complex construct that is almost impossible to create using traditional cloning methods.
Greg Foley:
With restriction cloning, we would typically pick eight colonies to screen by PCR and sequencing. But, now we sometimes just pick one or two, and can pretty much be sure that we're gonna get the right clone.
Yao Zhang:
Personally, I think the feature about NEBuilder that I like most is, it makes creating a point mutations much easier.
Tutorial: sgRNA Cas9 vector construction
NEB has developed a protocol to quickly and easily change the targeting of a single guide RNA in a sgRNA plasmid or to create an sgRNA plasmid library. The method uses a single DNA oligonucleotide, a restriction enzyme-digested plasmid and NEBuilder HiFi DNA assembly master mix.
Choose a target sequence or target sequences using a design tool of your choice. Design a single stranded DNA oligo containing a target sequence flanked by a partial promoter sequence and a scaffold RNA sequence.
In this animation, the U6 promoter is used. Prepare the DNA oligo. Assemble the reaction mix by combining DNA oligo, restriction enzyme linearized plasmid and water. Add NEBuilder HiFi DNA assembly master mix and incubate for one hour at 50 degrees Celsius.
Transform NEB 10 beta competent E. Coli with the assembled product. Spread out growth on plate with antibiotic and incubate overnight at 37 degrees Celsius. Pick colonies to grow and purify the plasmid DNA for sequencing. Some traditional methods require syntheses, phosphorylation, annealing and ligation of two oligos into a digested and dephosphorylated vector.
This new protocol, using a single DNA oligonucleotide with NEBuilder HiFi DNA assembly master mix is a simple and streamlined way to rapidly create specifically targeted cas9 sgRNA plasmids.
Deana Martin:
So, Tom, thanks so much for joining us today.
Tom Evans:
Thanks, it was a lot of fun.
Deana Martin:
Good. And as always, if you have any suggestions for future episodes, please let us know.
Related Videos
-
Introduction to NEBUILDER HIFI DNA ASSEMBLY® -

Construction of an sgRNA-Cas9 Expression Vector via an ssOligo Bridge -

NEBUILDER® HiFi: the next generation of DNA Assembly

