35-Fragment Golden Gate Assembly using BsmBI-v2 (NEB #E1602)
Overview
Golden Gate Assembly of 35 fragments using BsmBI-v2 restriction enzyme with T4 DNA Ligase and subsequent transformation into NEB 10-beta Competent E. coli (High Efficiency).
Materials
- NEBridge® Golden Gate Assembly Kit (BsmBI-v2) (NEB #E1602)
- pGGAselect Destination Plasmid*
- NEB 10-beta Competent E. coli (NEB #C3019)
- NEB 10-beta/Stable Outgrowth Medium (NEB #B9035)
- LB Agar plates with chloramphenicol
*included in NEBridge® Golden Gate Assembly Kit (BsmBI-v2) (NEB #E1602)
Reaction set-up
Set up assembly reactions as follows:
| REAGENT | ASSEMBLY REACTION |
|---|---|
| pGGAselect Destination Plasmid(1)*, 75 ng/μl | 1 μl |
| Amplicon Inserts(2) | 3 nM(3) each amplicon (final concentration) |
| T4 DNA Ligase Buffer (NEB #B0202) (10X) | 2 μl |
| NEBridge® Golden Gate Enzyme Kit (BsmBI-v2) (NEB #E1602) | 2 μl |
| Nuclease-free H2O (NEB #B1500) | to 20 μl(4) |
* or user provided
(2) Amplicon inserts must possess 5 ́ flanking bases (6 recommended) and BsmBI restriction sites at both ends of the amplicon
and in the proper orientation.
(3) The NEBiocalculator® Tool (nebiocalculator.neb.com) can be used for molarity calculations.
(4) Can be increased to 25 μl volume if required due to DNA component volumes; add additional 0.5 μl T4 DNA Ligase Buffer
(10X).
Reaction assembly protocol
| SUGGESTED ASSEMBLY PROTOCOL |
|---|
| (42°C, 5 min → 16°C, 5 min) x 30(5) → 60°C, 5 min → 4°C(6) |
(6) Cool reaction to 4°C prior to transformation, or store completed assembly reactions at -20°C.
Transformation
- For each assembly, thaw a 50 µl tube of NEB 10-beta competent E. coli cells on ice for 5–10 min.
- Add 2 µl of the assembly reaction; gently mix by flicking the tube 4-5 times.
- Incubate on ice for 30 min.
- Heat shock at 42°C for 30 sec.
- Place back on ice for 5 min.
- Add 950 µl of room temperature NEB 10-beta/Stable Outgrowth Medium (NEB #B9035). Incubate at 37°C for 60 min., shaking vigorously (250 rpm) or using a rotation device.
Plating
- Warm LB agar plates containing chloramphenicol (for pGGAselect) at 37°C for 15 min.
- Mix the cells thoroughly by flicking the tube and inverting, then spread 100 µl outgrowth onto each plate.
- Incubate the plates overnight at 37°C, or 24 hrs at 30°C, or 48 hrs at 25°C.
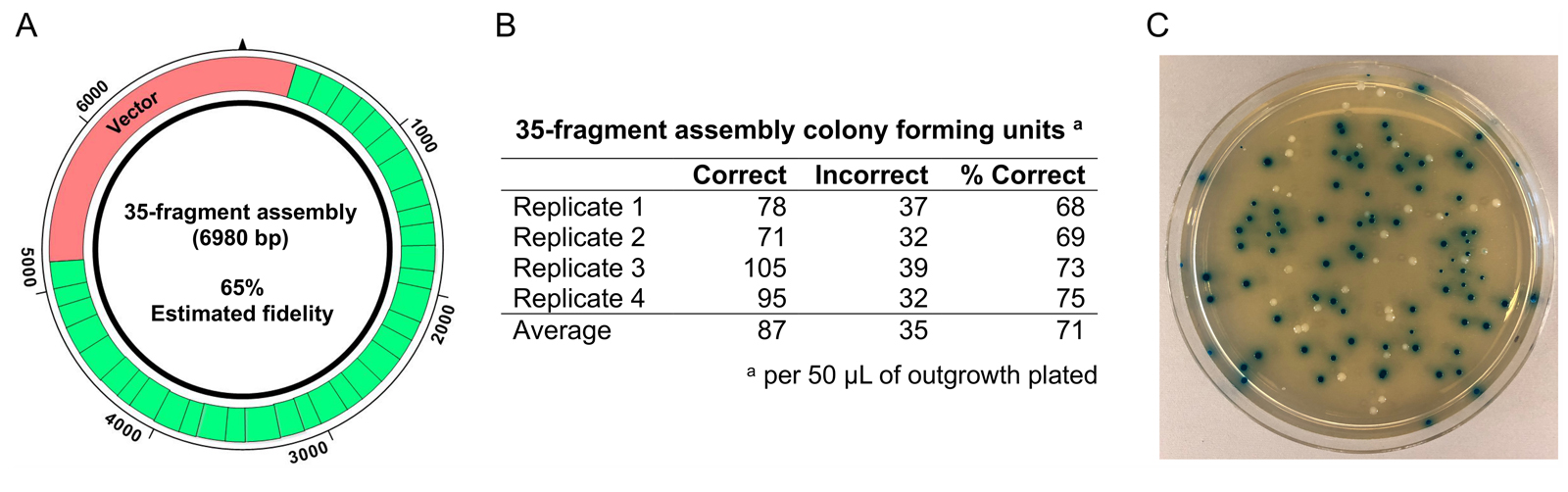
References
- NEBridge® Golden Gate Assembly Tool
- Ligase Fidelity Tools
- Golden Gate Assembly Resource Page
- Webinar: Fidelity and Bias in End-Joining Ligation: Enabling complex, multi-fragment Golden Gate DNA Assembly
- Pryor, J. M. et al. (2020) PLoS One, https://doi.org/10.1371/journal.pone.0238592


