
Library Preparation with FFPE DNA Samples
NEBNext Ultra II produces the highest yield libraries from a broad range of input amounts
A.| FFPE DNA | DNA INPUT (ng) | LIBRARY YIELDS IN ng | % MAPPED | % MAPPED IN PAIRS | % DUPLICATION | % CHIMERAS |
|---|---|---|---|---|---|---|
| Kidney Tumor | 17 | 132 | 91.5 | 96.1 | 0.48 | 3.0 |
| Lung Tumor | 20 | 232 | 90.1 | 94.9 | 0.42 | 4.1 |
| Liver Normal | 20 | 691 | 92.6 | 94.7 | 0.33 | 8.6 |
| Breast Tumor | 30 | 514 | 91.9 | 95.1 | 0.37 | 4.5 |
B.
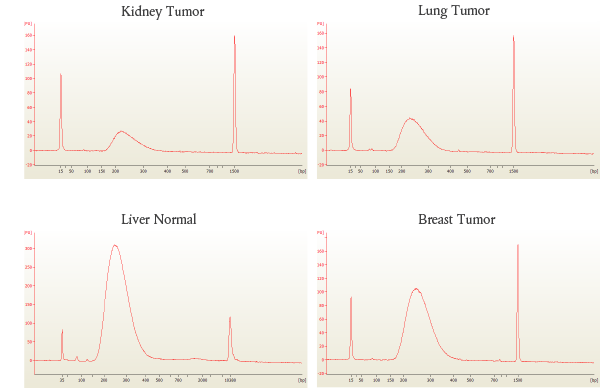
Libraries were prepared from 17–30 ng of human DNA extracted from the FFPE tissue samples listed, amplified using 10 cycles of PCR and sequenced on the Illumina MiSeq®. This data illustrates that NEBNext Ultra II enables high quality sequence data, even with low input amounts of FFPE DNA.
A: Reads were mapped to the GRCh37 reference genome using Bowtie 2.2.4.
% Mapped: The percentage of reads mapped to Human GRCh37 reference.
% Mapped in Pairs: The percentage of reads whose mate pair was also aligned to the reference.
% Duplication: The percentage of mapped sequence that is marked as duplicate.
% Chimeras: The percentage of reads that map outside of a maximum insert size or that have the two ends mapping to different chromosomes.
B: Bioanalyzer® traces of each library show high quality libraries with minimal adaptor-dimer.



