
Library Preparation for Target Enrichment Workflows
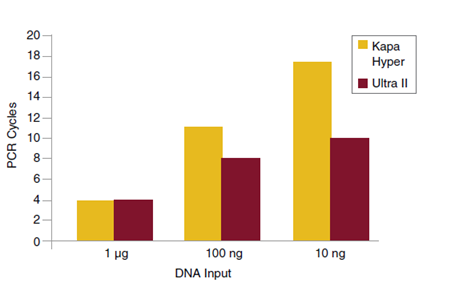
Libraries were prepared from Human NA19240 genomic DNA using NEBNext Ultra II and the input amounts shown. Yields were measured after each PCR cycle and the number of cycles required to generate at least 1 μg of amplified library determined. Cycle numbers for Kapa™ Hyper were obtained from Kapa Biosystems website and plotted alongside the cycle numbers obtained experimentally for Ultra II.
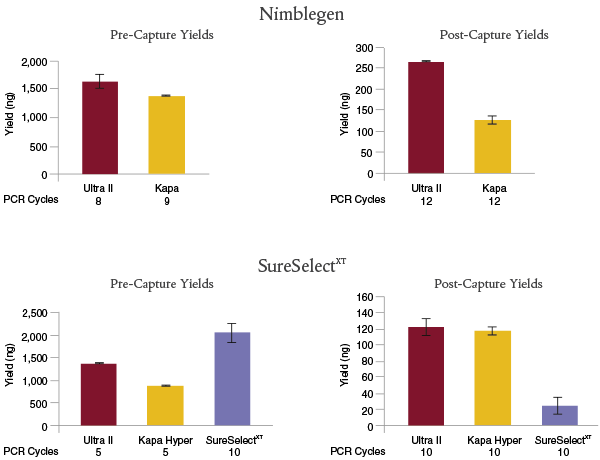
For Nimblegen® enrichment, libraries were prepared with 100 ng of Human HG02922 genomic DNA, using NEBNext Ultra II with the NEBNext Adaptor and index primers, or the Kapa Library Preparation Kit Illumina Platforms with the SeqCap® Adapter Kit (Roche). Target enrichment was performed with NimbleGen SeqCap Human Exome v3.
For SureSelect XT enrichment, libraries were prepared with 200 ng of Human HG02922 genomic DNA, using NEBNext Ultra II or Kapa Hyper with the NEBNext Adaptor and index primers, or the SureSelect XT Reagent Kit, Illumina (ILM) platforms with Herculase® II Fusion DNA Polymerase. Target enrichment was performed with SureSelect® Human All Exon V6.
Manufacturers’ recommendations were followed, including use of the recommended number of PCR cycles for pre- and post-capture. NEBNext Ultra II libraries resulted in the highest post-capture yields for both enrichment workflows.
| SAMPLE 1 | SAMPLE 2 | |||
|---|---|---|---|---|
| Library Prep Kit | Ultra II | Kapa | Ultra II |
Kapa |
| Total Reads | 81 M | 81 M |
81 M |
81 M |
| % Duplication | 9.2 | 14.7 | 9.7 | 12.4 |
| % Selected Base | 90.3 | 89.0 | 89.7 | 89.0 |
| Mean Target Coverage (x) | 54 | 50 | 53 | 51 |
| % Zero Coverage | 0.37 | 0.37 | 0.10 | 0.11 |
| Fold 80 Base Penalty | 2.56 | 2.77 | 2.65 | 2.81 |
| HS Library Size (molecules) | 167.9 M |
98.2 M |
156.1 M |
117.7 M |
Pre-capture libraries were prepared with 100 ng of Human HG00096 (Sample 1) and HG02922 (Sample 2) genomic DNA, using NEBNext Ultra II with the NEBNext Adaptor and index primers, or the Kapa Library Preparation Kit Illumina Platforms with the SeqCap Adapter Kit (Roche). Hybrid selection of the human exome was performed with 1 μg of each library and SeqCap human exome v3 following the NimbleGen SeqCap protocol. Post-capture amplification was conducted with NEBNext Ultra II Q5 Hot Start Master Mix for Ultra II libraries, and Kapa HiFi Ready Mix for Kapa librar ies. Libraries were pooled and sequenced on an Illumina NextSeq 500 instrument. Sequencing reads were mapped to human GRCh37 reference and analyzed using Picard HS Metrics tool.
% Duplication: The percentage of mapped sequence that is marked as duplicate.
% Selected Base: On+Near Bait Bases/PF Bases Aligned.
Mean Target Coverage: The mean coverage of targets that received at least coverage depth = 2 at one base.
% Zero Coverage: The number of targets that did not reach coverage=2 over any base.
Fold 80 Base Penalty: The fold over-coverage necessary to raise 80% of bases in “non-zero-coverage” targets to the mean coverage level in those targets.
HS Library Size: The estimated number of unique molecules in the library.
The NEBNext Ultra II Kit leads to lower duplication rate, higher percentage of selected base, lower Fold 80 Base Penalty, and significantly larger HS library size, suggesting a higher degree of uniformity of NEBNext Ultra II libraries.



