
Four alternative and powerful cloning applications of NEBuilder® HiFi DNA assembly
Posted on Monday, January 8, 2024
By
Topic: Tips for the lab, What is Trending in Science
The versatility of DNA manipulation techniques is crucial when addressing the wide variety of challenges faced by researchers. NEBuilder HiFi DNA Assembly (NEBuilder HiFi), typically known for facilitating seamless DNA fragment assembly, has several alternative yet highly effective applications. This article delves into four of these useful applications: bridging double-stranded DNA (dsDNA) fragments with a single-stranded (ss) oligonucleotide (oligo), annealed oligo assembly, mismatch repair, and multisite-directed mutagenesis. These techniques demonstrate NEBuilder HiFi's diverse utility in genetic engineering.

1. Bridging two dsDNA fragments with a ss oligo
Practical application
This alternative NEBuilder HiFi method has two primary uses:
(a) Adding small insertions, such as restriction sites or connecting two DNA fragments
(b) Constructing sgRNA libraries for CRISPR applications
(a) Adding small insertions
Overview
A bridging oligo can be customized to include additional sequences, offering flexibility for various genetic modifications. This technique is particularly useful for rapid and precise changes in plasmids.
Methodology
The process involves designing a ss oligo, typically around 60 base pairs, with 20-30 base pairs at each end, complementary to the DNA fragments being connected. The exonuclease activity in the NEBuilder HiFi mixture creates a 3´ overhang on the dsDNA, allowing the ss oligo to anneal. Subsequently, the high-fidelity DNA polymerase fills in the gaps, and DNA ligase seals the nicks.
This method is advantageous for sequences under 120 base pairs, where double-stranded fragments might be susceptible to degradation by the exonuclease before successful annealing.
Additional resources
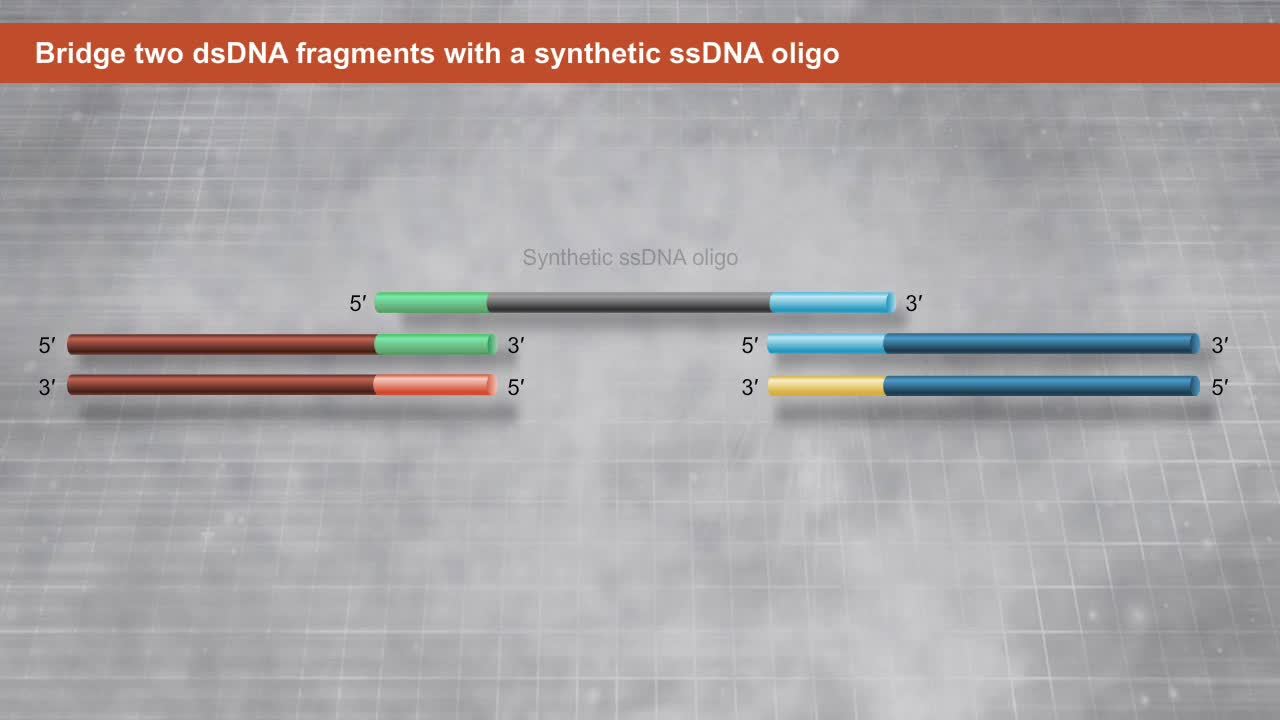
(b) Constructing sgRNA libraries for CRISPR applications
Overview
Creating plasmid libraries with variations in short sequences, such as those needed for CRISPR-based gene editing, is more feasible using ss oligos. Compared to the complexity of generating a library with dsDNA fragments for this application, ss oligos offer a streamlined approach.
Methodology
Researchers can order ss oligos as a pooled collection, each with specific overlaps for the sgRNA/Cas9 expression vector ends. NEBuilder facilitates this process as described above. It is notably efficient for generating varied sequences of about 15 base pairs over a short span. Transformation into bacteria results in colonies, each harboring a plasmid with a unique insert, thus efficiently generating a comprehensive library.
This method circumvents the challenges of assembling libraries using dsDNA fragments, which would require individual synthesis of each variant.
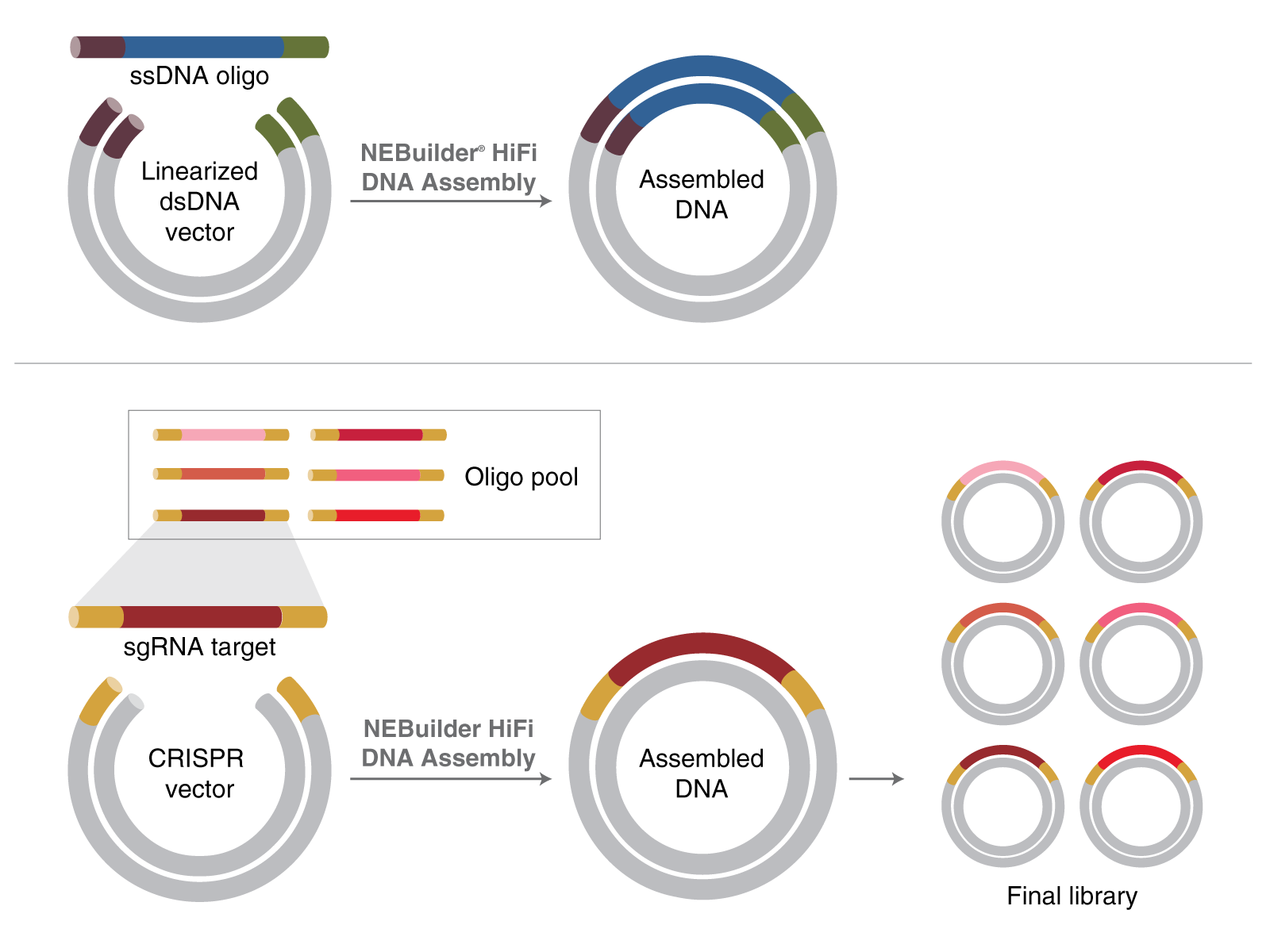
Top image: Adding small insertions. Bottom image: Constructing sgRNA libraries for CRISPR applications
Additional resources
Application note: Construction of an sgRNA-Cas9 expression vector via single-stranded DNA oligo bridging of double-stranded DNA fragments
2. Annealed oligo assembly
Practical application
This application provides an alternative to using synthesized dsDNA, such as gBlocks™.
Overview
The annealed oligo assembly technique, leveraging NEBuilder HiFi, provides a strategic approach for generating longer DNA sequences from short oligonucleotides. It stands out as an economical and reliable solution for assembling sequences where traditional methods fall short or when initial templates are unavailable.
Methodology
In this approach, short, overlapping ss oligonucleotides are annealed to form a longer, nicked dsDNA fragment. This is followed by an assembly reaction with a linearized vector and the NEBuilder HiFi DNA Assembly Master Mix.
This method is particularly useful for fragments that are challenging to amplify via PCR. It is cost-effective because large synthetic oligos are expensive to purchase. Rather than ordering a gBlock or attempting to use a long ss DNA, this method offers a viable alternative, especially in scenarios where no template is available for the desired insert.
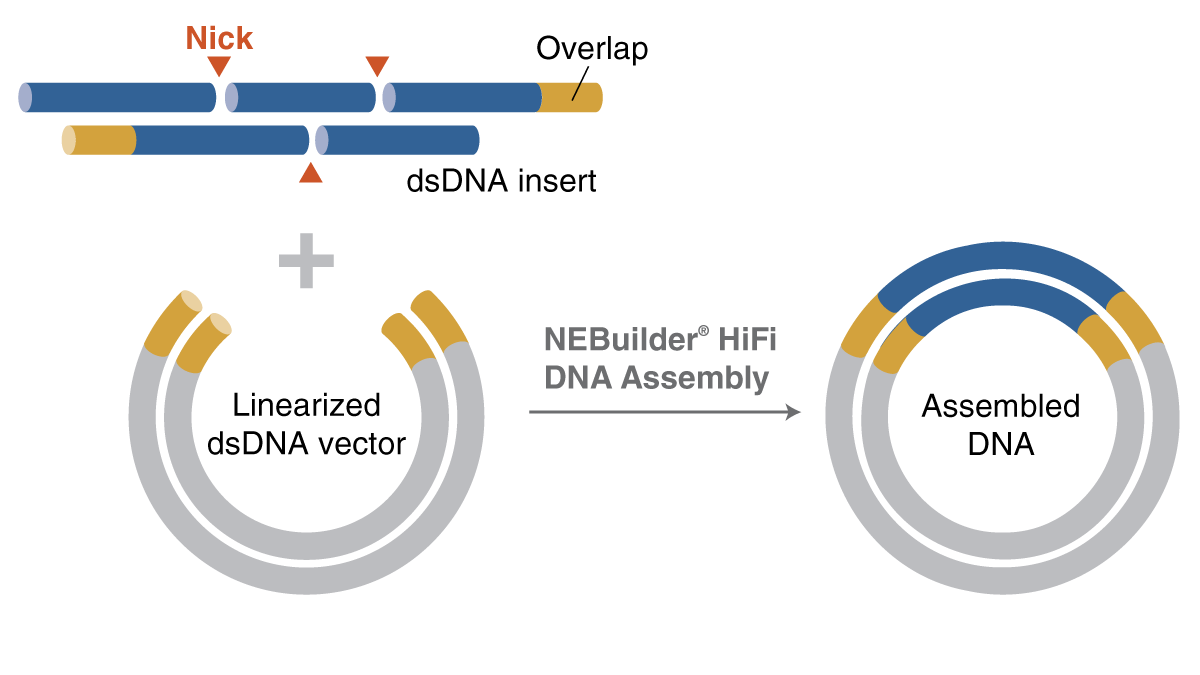
NEBuilder HiFi uses short ss oligonucleotides with overlapping regions to create a dsDNA fragment as an alternative to using synthesized dsDNA, such as gBlocks
Additional resources
3. Mismatch repair
Practical application
Removal of unwanted sequences, such as restriction sites, when creating genetic constructs.
Overview
NEBuilder HiFi excels in removing 3´ - and 5´ -end mismatch sequences during fragment assembly. This is particularly advantageous in scenarios where there are non-homologous sequences at the 3´ ends (beyond the regions of homology) that are unwanted in the final assembly product.
Methodology
The exonuclease removes bases from the 5´ end during fragment assembly, generating a 3´ overhang. If there are extra bases on the 3´ end that are not wanted in the final assembly product, NEBuilder HiFi is capable of efficiently removing 3´ end mismatches, up to 10 base pairs, so that they can be efficiently joined together.
When the homologous overlaps anneal during the assembly process, and there is a mismatch at the 3´ end, the proofreading DNA polymerase removes the extra bases, fills the gap, and DNA ligase seals the remaining nick.
Unlike other cloning methods, NEBuilder HiFi allows you to digest your DNA fragment with restriction enzymes and conveniently assemble it at any position.
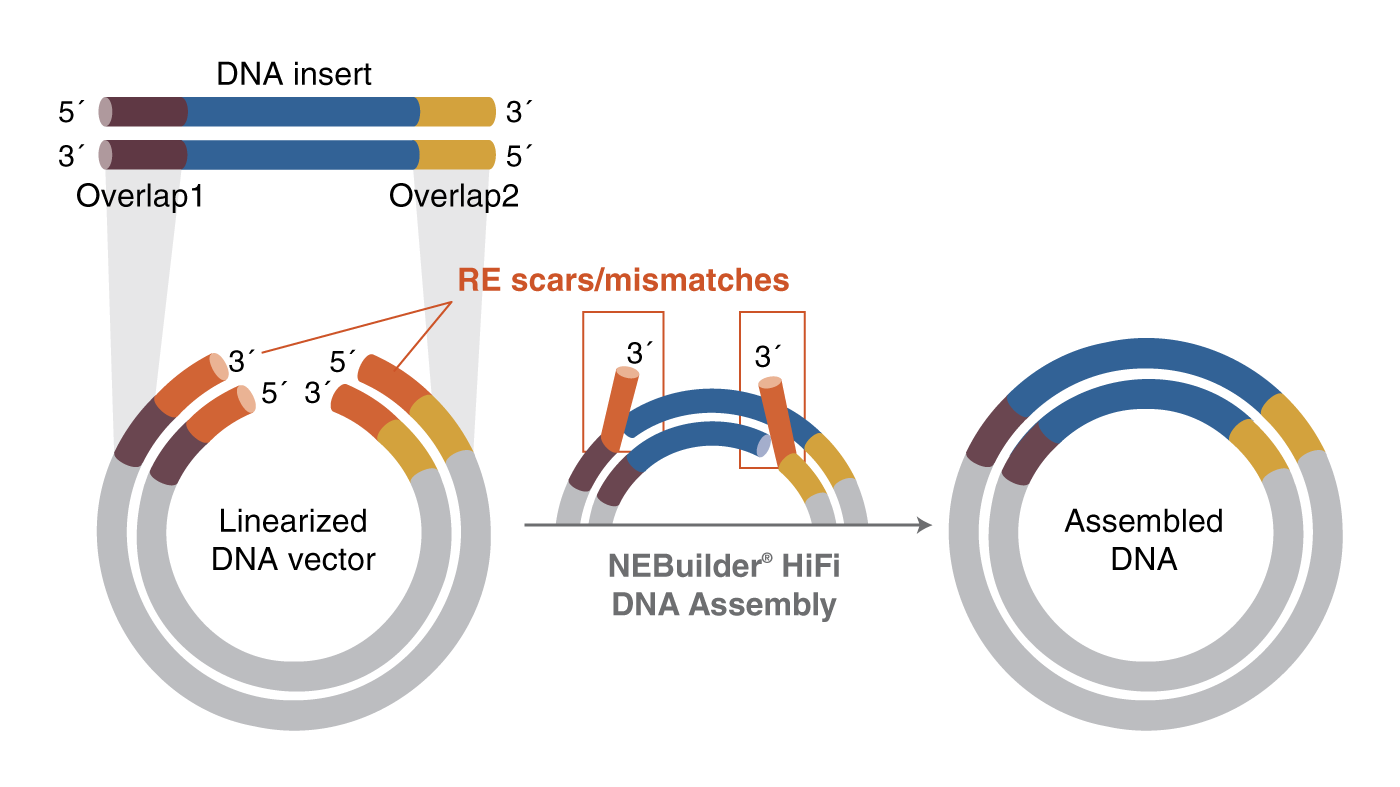
NEBuilder HiFi can remove unwanted sequences, such as restriction sites when creating genetic constructs
Additional resources

4. Multi site-directed mutagenesis
Practical application
NEBuilder HiFi can incorporate multiple mutations spaced far apart in a DNA sequence in a single reaction.
Overview
Traditional site-directed mutagenesis (SDM) methods, like whole plasmid SDM or PCR-site-directed mutagenesis, are adept at introducing mutations within a confined area. However, they falter when mutations are required at multiple sites that are not in close proximity, often becoming time-consuming, multistep workflows.
NEBuilder HiFi DNA Assembly Master Mix addresses this challenge by enabling the introduction of multiple mutations, regardless of their spacing within the DNA sequence. This capability is particularly beneficial when mutations are desired at intervals of several hundred base pairs within a gene or across different genes within the same plasmid.
Methodology
The NEBuilder HiFi method uses complementary flanking primers containing the desired mutations to align multiple PCR products or synthetic oligos with overlapping ends. These fragments are then assembled into a vector in a single-step reaction facilitated by the coordinated action of the three enzymes in the master mix. This process simplifies SDM and eliminates the need for phosphorylated primers, reducing cost and time.

NEBuilder HiFi can perform multi-site mutagenesis for diverse multi-site mutant library creation and screening
Additional resources
Application note: Improved methods for Site-Directed Mutagenesis using NEBuilder® HiFi DNA Assembly Master Mix
 |
Want to try NEBuilder HiFi DNA Assembly?Learn more |
NEB will not rent, sell or otherwise transfer your data to a third party for monetary consideration. See our Privacy Policy for details. View our Community Guidelines.
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. See www.neb.com/trademarks. The use of these products may require you to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
Don’t miss out on our latest NEBinspired blog releases!
- Sign up to receive our e-newsletter
- Download your favorite feed reader and subscribe to our RSS feed
Be a part of NEBinspired! Submit your idea to have it featured in our blog.


