.jpg?rev=3c161d1a378143bab628080a242e8d9d&sc_lang=en-us&h=520&w=2400&la=en-US&hash=6726DEEFCAB8BD01E1EC2F7C7002A252)
E5hmC-seq™ is a Sequencing Method for Direct 5hmC Detection that Leverages Improved Data Quality from Enzymatic Conversion
Posted on Friday, March 1, 2024
By
Topic: Tips for the lab
.png?sc_lang=en&rev=0f9f4f656da745dfb11cfba7548e8106&hash=C7E354AF4FA10B40ECE0C78F457FA359)
At NEB we get excited when a new method opens new avenues for research. Up until now, demonstrating the significance of 5-hydroxymethylcytosine (5hmC) has been impaired by a lack of robust methods to directly detect this modification at single-base resolution. NEBNext® Enzymatic 5hmC-seq (E5hmC-seq™) is a new method that removes that obstacle. As discussed in this interview with NEB scientist Dan Evanich, anticipation is high for the discoveries that will emerge from analyzing data using E5hmC-seq.
1. Why are epigenomics investigators so keen to map DNA hydroxymethylation?
Even though it’s still the subject of intense research, we know a lot about 5-methylcytosine (5mC), methylation at the fifth position of a cytosine base, which is the non-oxidized version of 5hmC. We have a good foundational understanding of 5mC dynamics. More recently, the field has become really interested in, and gained an appreciation for, the role that 5hmC may play in a variety of different systems. We know so much less about what 5hmC is doing and researchers are very interested in being able to map the distribution and abundance of this modification. They want to understand what its biological role is. We’ve only scratched the surface with our understanding of 5hmC dynamics. We’re excited that this new method will enable researchers to make a lot of interesting discoveries.
2. What are some of the common barriers scientists have faced sequencing genomes for 5hmC, up until now?
The top issue is that 5hmC is not very abundant in many different sample types. In some cases, it's not there at all. Levels of 5hmC tend to be intermediate. Methylation, and this is a bit of a generalization, but 5mC tends to be at 0 or 100% - a site is either fully methylated or fully unmethylated. Whereas 5hmC tends to exist in this intermediate range. It’s more dynamic. Some of the existing methods don’t offer the base level of information which I think is critical when you're trying to make certain biological assessments, like understanding what the level of hydroxymethylation is at specific sites and what they do. Another issue is that some other methods require you to prepare multiple libraries. For instance, you have a bisulfite library and another library type that detects only 5mC. You end up needing to perform this subtractive analysis to infer levels of 5hmC, rather than detecting 5hmC directly. Besides that, bisulfite-based conversion methods obviously damage DNA which leads to biased coverage of the genome. This really reduces the overall quality of data.
3. How does the NEBNext® Enzymatic 5hmC-seq method address those challenges?
The enzymatic reactions are really the key. It’s a two-step process and you need both robust protection and deamination for this method to work. We spent a lot of time making sure those enzymatic reactions were very efficient. We see excellent protection of 5hmC and highly efficient deamination of the non hydroxymethylated sites. We wanted to ensure that investigators aren't restricted by the amount of material that they have. As we were developing the kit for this method (NEB# E3350), we optimized it to be compatible with a wide input range, between 0.1–200 ng. That makes it a robust solution that scientists can go in with confidently. Internal controls are included in the kits to give investigators a way to validate the enzymatic reactions are performing as expected. Even though everything is already optimized, it’s nice to have that additional confirmation that everything is working. Enabling direct detection of 5hmC is a real strength. The readout is 5hmC, rather than an inference based on a reversed readout. When you're sequencing multiple libraries to do a subtractive analysis to infer the modification that you're interested in, that compounds error. Using the E5hmC-sequencing method avoids that risk. Again, the fact that we don't have a bisulfite conversion step also vastly improves the data quality. Not only is there a direct assessment of 5hmC, but we have all of the advantages of enzymatic conversion from library yields to even GC coverage from a data quality perspective.
.png?sc_lang=en&rev=3885650ee29243d68135568eeff3c076&hash=998FA26B5674BDBE6FABA91530350297)
Figure 1: 5hmC detected by E5hmC-seq in human brain gDNA is consistent across inputs. 200-0.1ng of human brain genomic DNA was sheared to 350bp (Covaris® ME220) and E5hmC-seq libraries were prepared and sequenced on an Illumina® NovaSeq® 6000 (2 x 150 bases). Approximately 1.9 billion (200ng, 10ng and 1ng) reads for each library were aligned to a composite human T2T, lambda and T4 reference genome using bwa-meth, and methylation information was extracted from the alignments using MethylDackel. Values shown are the average of two technical replicates and error bars show standard deviation. Detected 5hmC levels are similar between all inputs in the CpG, CHH and CHG contexts.
4. How does E5hmC-seq work to detect 5hmC only?
The NEBNext® E5hmC-seq method (NEB #E3350) is similar to NEBNext® Enzymatic Methyl-seq (EM-seq) (NEB #E7120) since it relies on a protection and deamination scheme. The first step in the full kit is preparation of your library. This entails fragmentation, end repair and dA-tailing and ligation to sequencing adaptors. Then 5hmC is protected specifically using T4 Phage β-glucosyltransferase (T4-BGT), that glucosylates 5hmC only, but doesn't touch 5mC or unmethylated cytosine. In the second reaction, deamination of 5mC and unmethylated cytosine is carried out by APOBEC. The deaminase doesn't touch the glucosylated and protected 5hmC moiety. In the end you have 5hmC sites being sequenced as C and your cytosine and methylated cytosine sites being sequenced as T.
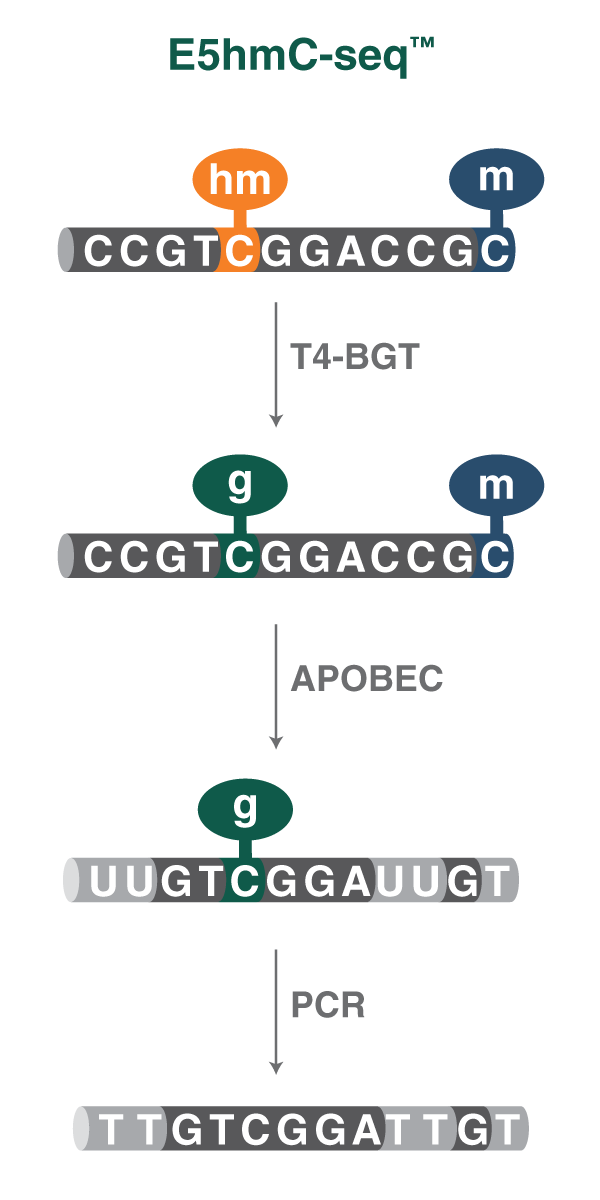
To enable specific 5hmC detection, 5hmC is first glucosylated using T4-BGT. 5mC and unmodified cytosine are then deaminated by APOBEC to thymine and uracil, respectively, while the protected 5hmC is not converted. During Illumina sequencing, 5hmCs are represented as cytosine, while unmethylated cytosine and 5mCs are represented as thymine.
5. Can raw data from E5hmC-seq be used in WGBS bioinformatics pipelines?
The way that the conversion scheme works, the modified base that you're interested in is sequenced as C and everything you're not interested in is sequenced as T. This is the same as you would see for EM-seq or bisulfite converted DNA so you can use existing informatic pipelines that have been developed for those methods. The only difference is that the methylation signal you’re getting at the end is specifically 5hmC.
6. Can you share some compatible applications that have clinical diagnostics potential?
As I mentioned, there is increasing interest in 5hmC, and we’ve really just begun to scratch the surface. There haven’t been robust methods to look at 5hmC. I think the first step is that people are going to use this method, explore hmC distribution in their samples of interest and identify what is clinically actionable. To that end we’ve developed this method to be compatible with low inputs, as well as different clinically relevant sample types such as cell free DNA (cfDNA). Our approach has been to enable the scientific community to reliably process samples with the types of constraints they would be operating under for diagnostics development.
7. Are researchers also decoding 5hmC in non-mammalian models?
Most of the non-human work has been in mouse and increasingly people are starting to explore different organisms. It’s just as critical in these cases to have a robust solution. That’s a force that will drive this research area forward.
8. What do you anticipate the impact of this method could be?
Investigators have all this interesting data from EM-seq and bisulfite sequencing that contains a composite of methylated cytosine and hydroxymethylated cytosine from so many sample types. In some cases, it may be that a proportion of that might be hmC that wasn't appreciated before because you couldn’t differentiate it. Not only is this a very interesting new data type, but by leveraging all the benefits of the enzymatic conversion process, with respect to yields and data quality, it offers a clearer view on 5hmC.
As a scientist, it's been fun to be involved in the development of this technique, because I've learned so much about 5hmC myself, which is something that the research community itself is still trying to understand.
NEB will not rent, sell or otherwise transfer your data to a third party for monetary consideration. See our Privacy Policy for details. View our Community Guidelines.
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. See www.neb.com/trademarks. The use of these products may require you to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
Don’t miss out on our latest NEBinspired blog releases!
- Sign up to receive our e-newsletter
- Download your favorite feed reader and subscribe to our RSS feed
Be a part of NEBinspired! Submit your idea to have it featured in our blog.


