What are endonucleases and their applications?
Script
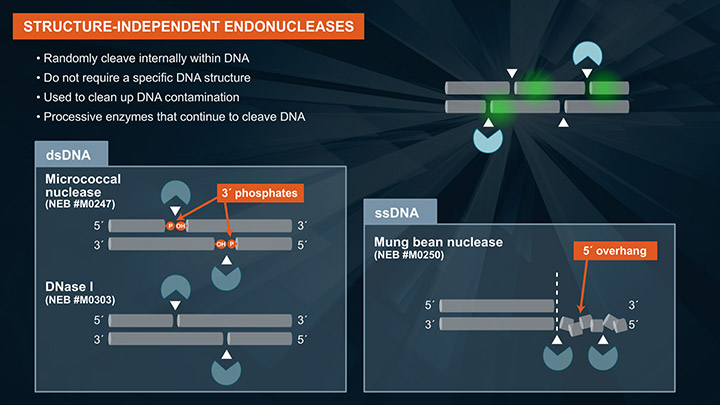
Endonucleases cleave DNA internally, and do not require free DNA ends for activity. This is in contrast to exonucleases, which require and act on free DNA ends. In vivo, the primary role of endonucleases is to repair damaged DNA. Many of these enzymes have been adapted into workflows such as gene assembly and directional RNA-seq. Here, we will be highlighting two types of endonucleases: structure-independent and structure-dependent. Structure-independent endos randomly cleave internal phosphodiester bonds, and are named such because they do not require a specific DNA structure for activity. They act on canonical single and double stranded DNA. Structure-independent endos are primarily used to clean-up DNA contamination. These are processive enzymes, and once activity has been initiated, the endo will continue to cleave DNA. Examples of structure-independent endos active on double-stranded DNA are micrococcal nuclease and DNase I. An example of a structure-independent endo active on single-stranded DNA is mung bean nuclease. Structure-dependent endos, which include wide variety of DNA repair enzymes, require a specific DNA structure or damaged base for activity, and will cleave an internal phosphodiester bond at that structure or damaged base. Apurinic or apyrimidinic sites, also known AP or abasic sites are common and occur spontaneously in DNA when a base is lost. This can be detrimental to the cell. An example of an endonuclease that will act on an AP sites is Endo IV. It catalyzes the cleavage of DNA phosphodiester backbone at AP sites via hydrolysis, leaving a 1 nucleotide gap with 3´-hydroxyl and 5´-deoxyribose phosphate (dRP) termini. 8-oxoguanine, more commonly known as 8-oxoG, is a common DNA error that results from oxidizing guanine. This is also detrimental to the cell. An example of an DNA repair enzyme that removes this type of error is Fpg. Fpg is a bifunctional DNA glycosylase with DNA N-glycosylase and AP lyase activities. N-glycosylase activity releases damaged purines, including 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) and 8-oxo-7,8-dihydroguanine (8-oxoG), generating an AP site. The AP lyase activity of Fpg cleaves an AP site, via beta and delta-elimination, creating a 1 nucleotide DNA gap with 5-prime and 3-prime phosphate termini. Some enzymes act only as glycosylases, that is, they only recognize and remove the damaged base but do not specifically cleave the DNA backbone. An example of this type of enzyme is UDG, which recognizes deoxyuridine, or dU, and removes it and leaves an AP site. To cleave the DNA backbone, another enzyme such as Endonuclease VIII is required. USER Enzyme contains a mixture of these two enzymes so that the damage is completely removed, and the AP site is cleaved. Aside from DNA repair enzymes, there are also structure-specific endonucleases that act on DNA secondary structure. Thermostable FEN1, for example, recognizes a specific FLAP structure, and cleaves this flap leaving a 5-prime phosphate, which can then be ligated with a DNA ligase.
For more information regarding NEB endonucleases, please visit the following resources at www.neb.com.

