Addressing Challenges in Microbiome DNA Analysis
Among the very many “-omes” now studied and discussed (1), microbiomes have received increasing attention in recent months, from
both scientists and the general public. Used to describe the communities of microorganisms and their genes in a particular environment,
including a body or part of a body, “microbiome” is becoming an increasingly common term in everyday language. One challenge in
microbiome genome analysis is addressing the presence of host DNA in samples. As such, improved methods for solving this problem
are needed.
Fiona Stewart, Ph.D. and Erbay Yigit, Ph.D., New England Biolabs, Inc.
Introduction
A wealth of information about the composition
of, and interactions between, the constituent microbes
of a microbiome can provide insight into
both the function and dysfunction of the host
organism, as well as the host-microbiome unit as
a whole. In particular, the relationships amongst
and between resident microbes (bacteria, archaea and fungi) and their hosts have recently become
the topic of fervent research; the number of microbiome
research publications has been steadily
increasing since 2003 (2). Such research has
demonstrated that the microbiome communities
of individuals are unique, as are the microbiome
communities of specific sites within an individual
(reviewed in 3). In humans, the number of microorganisms
present is estimated to exceed the
number of human cells by 10-fold (4). Studies
of the human microbiome (including the Human
Microbiome Project (HMP) [www.hmpdacc.org]
(5), and MetaHIT, the metagenomics of the intestinal
tract [www.metahit.eu] (6)) may be the best
known, and have led to the understanding that
the human microbiome may be critical to health
and disease.
Until relatively recently, the role of the microbiome
was unknown, and an organism’s microbial
load was considered to be potentially nothing
more than cellular “hitchhikers”, having little
impact on the organism’s functioning. Now, it is
understood that an organism’s microbiome can
influence many processes within the host organism.
Discoveries including the role of the microbiome
in conditions and disease states, such as
obesity, diabetes mellitus and cardiovascular disease
(reviewed in 7), have led to the potential for
development of microbiome-based diagnostic and
therapeutic tools. Additionally, the unique nature
of an individual’s microbiome has enabled matching
of skin-associated bacteria, on objects such
as a keyboard, to specific individuals, leading to
the potential for use in forensic applications (8). It
should be noted that microbiome research is not
limited to humans, and research into microbiomes
of non-human organisms is also increasing rapidly in environmental and agricultural areas
of research (9).
Although it is still not possible to isolate and culture
the vast majority of microorganisms (estimated
to be over 95%), analysis of total nucleic acid
from microbiome samples has enabled significant
advances in the field. Furthermore, advances in
sequencing technologies have enabled significant
progress in microbiome nucleic acid analysis.
Current Methods of Analysis
The majority of microbiome DNA studies to date have employed 16S analysis (Figure 1). This analysis method takes advantage of the 16S rRNA gene that is specific to prokaryotes and some of the archaea and is not found in eukaryotes. 16S rRNA genes from different species have significant homology, but the gene also includes hypervariable regions that are generally speciesspecific, and are determined by the microbial composition of the community. These characteristics enable the use of universal primer pairs to amplify 16S genes from many organisms in the same PCR reaction and then, through subsequent sequencing of the PCR products, the individual species represented can be identified.
Figure 1. Microbiome DNA Analysis Methods
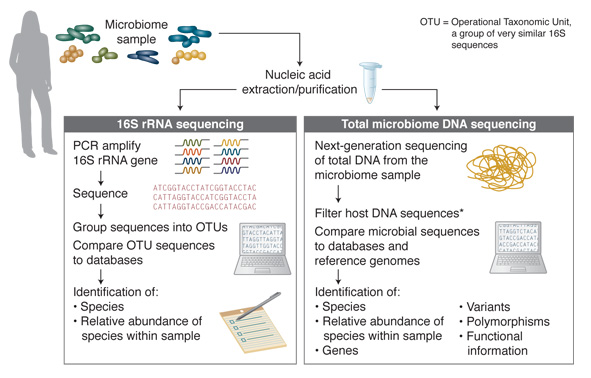
* For many samples, host DNA constitutes a high percentage of sequence reads. Removal of host DNA, and enrichment of microbial DNA substantially increases the percentage of sequence reads from the microbial sequences of interest.
While the 16S method is a fast and relatively inexpensive way to survey, at high throughput, the microbial organisms present within a sample, it provides very little information regarding function. Additionally, determining optimal PCR primers (for specific sample types and to distinguish between some species) can be challenging. In contrast, sequencing of the total DNA of a microbiome sample does not have these limitations and provides a more complex range of information. Through the identification of microbial sequences, genes, variants and polymorphisms, this method enables determination of information on microbiome species diversity and, also, putative functional information. Such sequencing-based studies have enabled the creation of many databases, including the Human Oral Microbiome Database (HOMD) [www.homd.org] (10). Approximately 700 prokaryotic species are present in the human oral cavity, and the stated goal of the HOMD database project is to provide taxonomic and genomic information on these species. Comparison of microbiome sample sequences to databases, such as HOMD, further enables discovery, including genes, pathways and their relative frequencies in the sample.
Overcoming Difficulties
with Microbiome Samples
Many microbiome samples are overwhelmed with
host DNA, and the HMP has reported especially
high levels of human DNA in soft tissue samples,
such as mid-vagina and throat samples. Saliva
samples also contain high levels of human DNA
(11). In contrast, although human DNA is generally
all but absent from fecal samples, some infections
can substantially increase the level of human
DNA in such samples, likely due to widespread
cell lysis during bacterial infection.
The presence of contaminating host genomic
DNA in a microbiome sample complicates the
genetic analysis of these samples. Since a single
human cell contains approximately 1,000 times
more DNA than a single bacterial cell (approximately
6 billion bp versus 4-5 million bp), even
a low level of human cell contamination within a
microbiome sample can substantially complicate
the sample processing and sequencing. As a result, in the case of total microbiome DNA sequencing
studies, only a small percentage of sequencing
reads from such samples pertain to the microbes
of interest, and therefore a large percentage of
sequencing reads (host) have to be discarded.
Consequently, obtaining sufficient sequence coverage
of the microbiome DNA can become costprohibitive
or even technically infeasible. Therefore,
methods to enrich microbiome DNA are
useful, and, in some cases, critical for sequencing
of the microbiome. However, until now, options
for such enrichment have been limited to selective
cell lysis, with the disadvantages of a requirement
for live cells, and low bacterial DNA recovery.
The NEBNext® Solution
The NEBNext Microbiome DNA Enrichment Kit
addresses this problem by providing a quick and
effective way to remove contaminating host DNA,
thereby enriching for microbiome DNA. The kit
exploits the different prevalences of CpG methylation
in the genomes of microbial and eukaryotic
organisms. Eukaryotic DNA, including human
DNA, is methylated at CpGs, while methylation
at CpG sites in microbial species is rare.
The NEBNext Microbiome DNA Enrichment Kit
uses a magnetic bead-based method to selectively
bind and remove CpG-methylated host DNA.
feature article continued…
The kit contains the MBD2-Fc protein, which is
composed of the methylated CpG-specific binding
protein MBD2, fused to the Fc fragment of
human IgG. The Fc fragment binds readily to
Protein A, enabling effective attachment to Protein
A-bound magnetic beads. The MBD2 domain
of this protein binds specifically and tightly
to CpG methylated DNA. Application of a magnetic
field then pulls out the CpG-methylated (eukaryotic)
DNA, leaving the non-CpG-methylated
(microbial) DNA in the supernatant.
Microbiome Enrichment of Human Saliva
Human saliva samples can be especially challenging, due to high levels of human genomic DNA and the poor-quality of the DNA itself. Despite these sample challenges, the data shown in Figure 2 demonstrates that substantial enrichment of microbiome DNA from saliva was achieved using the NEBNext Microbiome DNA Enrichment Kit.
Figure 2. Salivary Microbiome DNA Enrichment
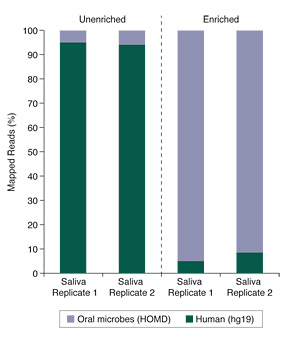
Figure 3. Microbiome Diversity is Retained After Enrichment with the NEBNext Microbiome DNA Enrichment Kit
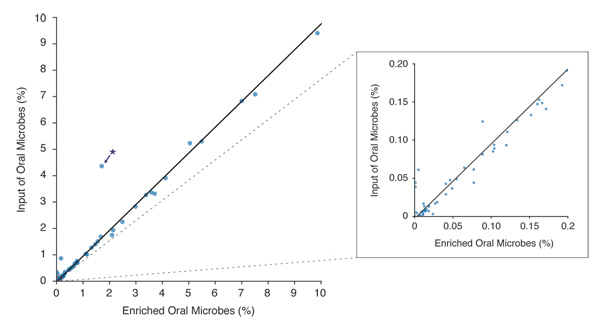
* Niesseria flavescens – This organism may have unusual methylation density, allowing it to bind the enriching beads at a low level. Other Niesseria species (N. mucosa, N. sicca and N. elognata) are represented, but do not exhibit this anomalous enrichment.
Conclusion
From forensic microbial “fingerprints” to disease-causing pathogens, microbiomes comprise a vast and varied microcosm with a surprising degree of influence over the health and function of the host organism. The potential for significant and exciting discoveries to be achieved with microbiome analysis is enormous, but will require improved tools and methods to make this a reality. As a step towards this goal, the NEBNext Microbiome DNA Enrichment Kit now makes it possible to substantially enrich a variety of sample types for non-host, microbial DNA, while retaining microbial diversity, and thereby improving the quality and cost-effectiveness of downstream analyses and data generation.
References
- Alphabetically ordered list of -omes and -omics (2013) Omics. org Retrieved on May 1, 2013, from www.omics.org.
- Jones, S. (2013) Nature Biotechnology, 31, 277.
- Morgan, X.C., et al. (2013) Trends in Genetics, 29, 51–58.
- Backhed, F., et al. (2005) Science, 307, 1915–1920.
- Peterson, J., et al. (2009) Genome Res. 19, 2317–2323.
- Qin, J., et al. (2010) Nature, 464, 59–65.
- Pflughoeft, K.J. and Versalovic J. (2012) Annu. Rev. of Pathol. 7, 99–122.
- Fierer N., et al. (2010) Proc. Natl. Acad. Sci. USA, 107, 6477–6481.
- Jansson, J.K. and Prosser, J. I. (2013) Nature, 494, 40–41.
- Chen, T., et al. (2010) The Human Oral Microbiome Database Retrieved on May 1, 2013, from www.homd.org.
- The Human Microbiome Project Consortium (2012) Nature, 486, 215–221.
- Langmead, B., et al. (2009). Genome Biol. 10(3), R25.

