Enzymes For Innovation
Products Available
| 5-hydroxymethyluridine DNA Kinase | M0659 | 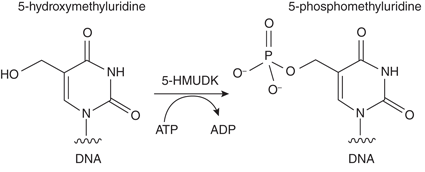 5-hydroxymethyluridine DNA Kinase (5-HMUDK) transfers the gamma phosphate from ATP to the hydroxymethyl moiety of 5-hydroxymethyluridine in polymeric DNA. |
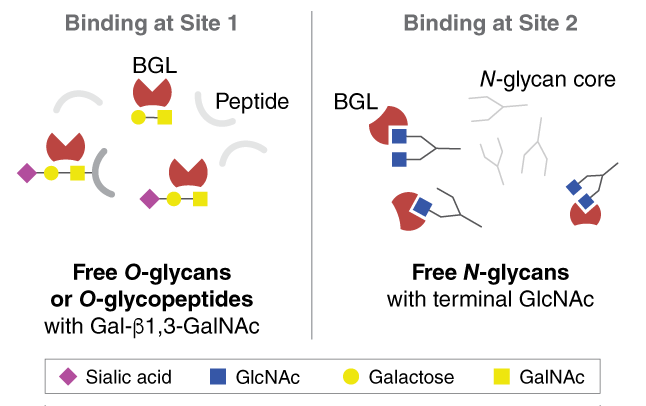
| Boletopsis grisea Lectin (BGL) |
P0867 |
Enables enrichment of GlcNAc-capped N-glycans or mucin type O-glycopeptides from complex samples in glycomics and glycoproteomics analytical workflows. |
| EcoGII Methyltransferase | M0603 |  EcoGII Methyltransferase is a non-specific methyltransferase that modifies adenine residues (N6) in any sequence context. |
| Endoglycoceramidase I (EGCase I) | P0773 | 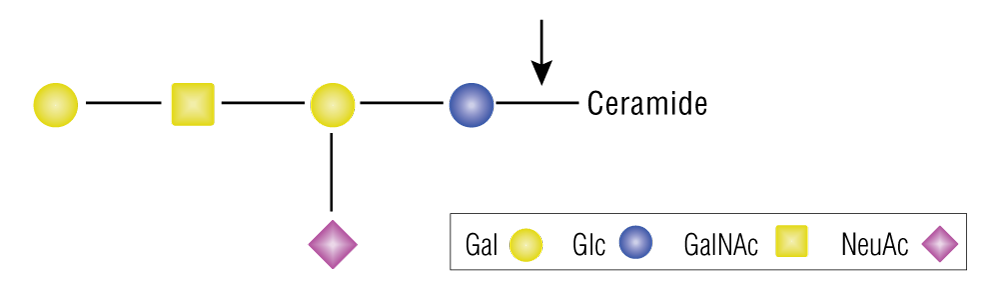 Catalyzes the hydrolysis of the β-glycosidic linkage between oligosaccharides and ceramides in various glycosphingolipids. |
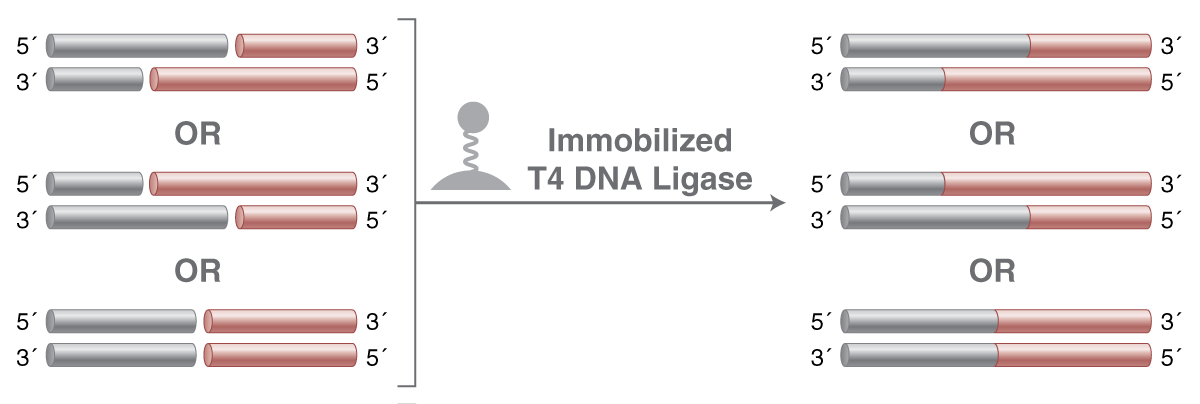
| Immobilized T4 DNA Ligase |
M0569 |
Immobilized T4 DNA Ligase is a slurry of magnetic beads coated with T4 DNA Ligase to produce a 10 mg/ml solution (50% glycerol) with an effective concentration of 60 cohesive end units (CEU) per microliter of slurry. Following a reaction, the enzyme can be removed using a magnet and can be re-used. |
| Msz Exonuclease I (MszExol) |
M0527 |  Catalyzes the removal of nucleotides from linear single-stranded DNA in the 3′ to 5′ direction with optimal activity between 45°C and 60°C. |
| Nb.BssSI | R0681 |  Generates a nick in the 3´ strand at the following recognition site: CACGAG (none/-1). |
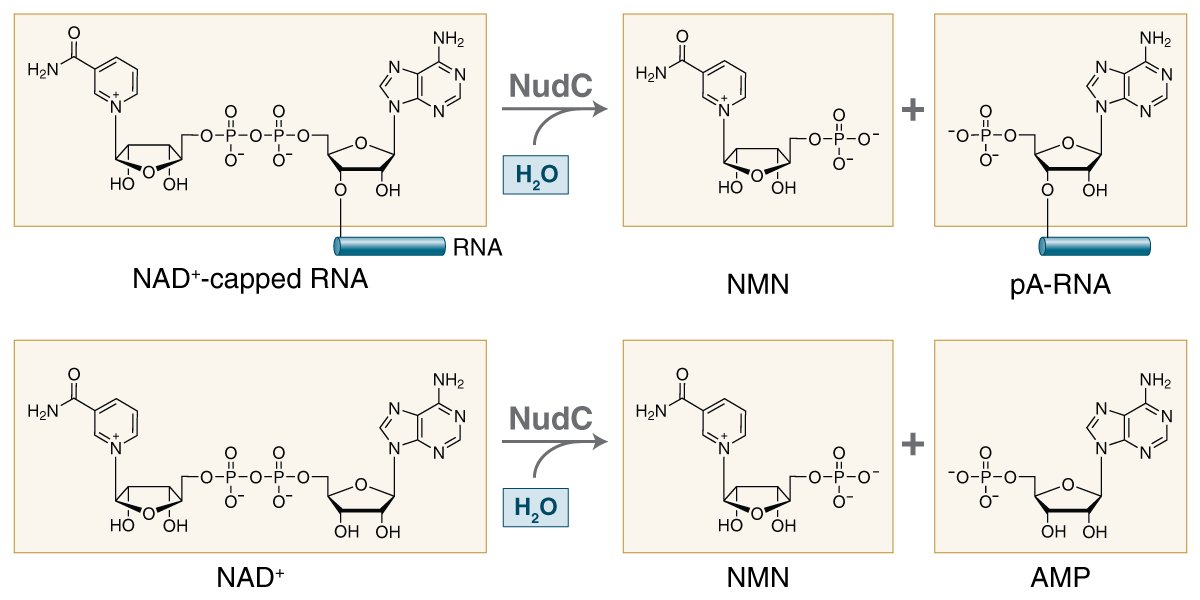
| NudC Pyrophosphatase | M0607 |
NudC is a NUDIX pyrophosphatase that efficiently hydrolyzes NAD+ - and NADH-capped RNA, generating a ligatible 5´ monophosphate on the RNA (NAD+ decapping or deNADding). |
| RtcB Ligase | M0458 | 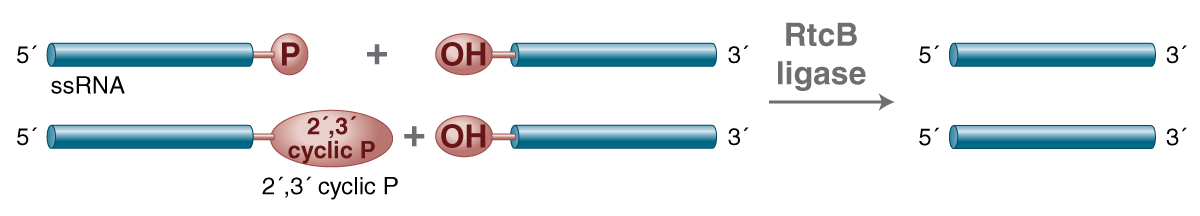 Joins single-stranded RNA with a 3´-phosphate or 2´,3´-cyclic phosphate to another RNA with a 5´ -hydroxyl. |
| Sce PUS1 |
M0526 |
Sce Pseudouridine Synthase I (Sce PUS1) converts Uridine to Pseudouridine in single-stranded RNA, with a preference for Uridines in single-stranded RNA regions over Uridines in double-stranded RNA. The optimal substrate is an unstructured RNA that is 15 nt long or longer. |
| TelN Protelomerase | M0651 |  Cuts dsDNA at a TelN recognition sequence (56 bp) and leaves covalently closed ends at the site of cleavage. |
| Thermostable FEN1 | M0645 |  Catalyzes the cleavage of 5´ DNA flaps from branched double-stranded DNA substrates. |
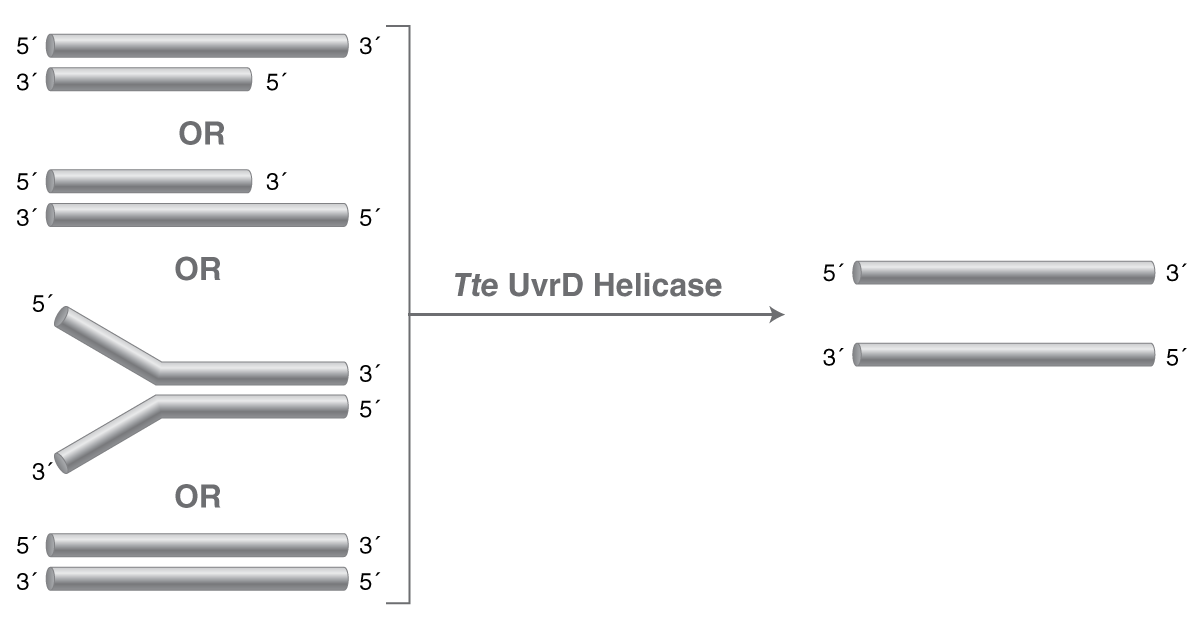
| Tte UvrD Helicase | M1202 |
Tte UvrD Helicase is a repair helicase capable of unwinding double-stranded DNA, without a requirement for a specific flap or overhang structure, from the thermophilic organism Thermoanaerobacter tengcongensis. |
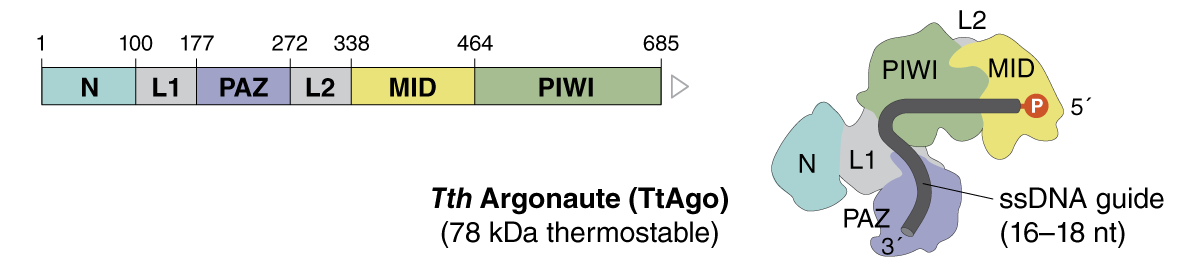
| Tth Argonaute | M0665 |
TtAgo is a prokaryotic argonaute that functions as a DNA-guided endonuclease when provided with a 16–18 nucleotide long 5´-phosphorylated single-stranded DNA oligonucleotide guide. |
If you are looking for an enzyme functionality that is not currently available, contact EnzymesForInnovation@neb.com.
Have you found a good application for one of the enzymes listed above? we would love to hear about it!
Choose Type:
A: Enzymes for Innovation (EFI) is a project initiated by NEB to provide unique enzymes to the scientific community in the hopes of enabling the discovery of new and innovative applications. These enzymes have interesting properties and unique specificities that are not commercially available elsewhere at the quality that you would expect from NEB. If you have an idea for an “Enzyme for Innovation” with a suggested application that may be useful, please email us at EnzymesForInnovation@neb.com. For more information, please view video.
Q: Can I submit an Enzyme for Innovation idea?
A: Yes! Please email EnzymesForInnovation@neb.com with ideas for new enzymes (please note enzymes should modify either DNA, RNA, or protein, or be useful in glycobiology applications).
Q: What quality controls are performed to validate Enzymes for Innovation?
A: Products designated as Enzymes for Innovation pass stringent quality controls, including removal of contaminants and characterization of enzyme stability.