QC Check and Size Selection using 6% PolyAcrylamide Gel (E7330)
- Purify the PCR amplified cDNA construct (100 μl) using a QIAQuick PCR
Purification Kit.
IMPORTANT: Before eluting the DNA from the column, centrifuge the column with the lid of the spin column open for 5 minutes at 13,200 rpm. Centrifugation with the lid open ensures that no ethanol remains during DNA elution. It is important to dry the spin column membrane of any residual ethanol that may interfere with the correct loading of the sample on the PAGE gel.
- Elute amplified DNA in 27.5 μl Nuclease-free Water.
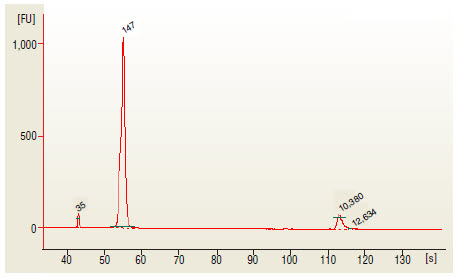
- Load 1 μl of the purified PCR reaction on the Bioanalyzer using a DNA 1000
chip according to the manufacturer's instructions (Figure 1).
- Mix the purified PCR product (25 μl) with 5 μl of Gel Loading Dye, Blue (6X).
Note: Vortex the Gel Loading Dye, Blue throughly to mix well before using.
- Load 5 μl of Quick-Load pBR322 DNA-MspI Digest in one well on the 6%
PAGE 10-well gel.
- Load two wells with 15 μl each of mixed amplified cDNA construct and
loading dye on the 6% PAGE 10-well gel.
- Run the gel for 1 hour at 120 V or until the blue dye reaches the bottom of
the gel. Do not let the blue dye exit the gel.
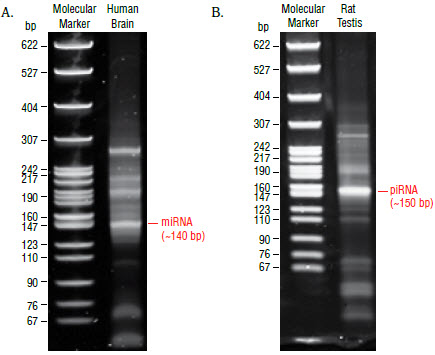
- Remove the gel from the apparatus and stain the gel with SYBR Gold
nucleic acid gel stain in a clean container for 2–3 minutes and view the gel
on a UV transiluminator (Figure 2).
- The 140 and 150 nucleotide bands correspond to adapter-ligated
constructs derived from the 21 and 30 nucleotide RNA fragments,
respectively. For miRNAs, isolate the bands corresponding to ~140 bp.
For piRNAs, isolate the band corresponding to ~150 bp.
- Place the two gel slices from the same sample in one 1.5 ml tube and
crush the gel slices with the RNase-free Disposable Pellet Pestles and
then soak in 250 μl DNA Gel Elution buffer (1X).
- Rotate end-to-end for at least 2 hours at room temperature.
- Transfer the eluate and the gel debris to the top of a gel filtration column.
- Centrifuge the filter for 2 min at > 13,200 rpm.
- Recover eluate and add 1 μl Linear Acrylamide, 25 μl 3M sodium acetate,
pH 5.5 and 750 μl of 100% ethanol.
- Vortex well.
- Precipitate in a dry ice/methanol bath or at –80°C for at least 30 minutes.
- Spin in a microcentrifuge @ > 14,000 x g for 30 minutes at 4°C.
- Remove the supernatant taking care not to disturb the pellet.
- Wash the pellet with 80% ethanol by vortexing vigorously.
- Spin in a microcentrifuge @ > 14,000 x g for 30 minutes at 4°C.
- Air dry pellet for up to 10 minutes at room temperature to remove residual
ethanol.
- Resuspend pellet in 12 μl TE Buffer.
- Load 1 μl of the size selected purified library on a 2100 Bioanalyzer using
a DNA 1000 or High Sensitivity DNA chip according to the manufacturer's
instructions (Figure 3).
- Check the size, purity, and concentration of the sample.
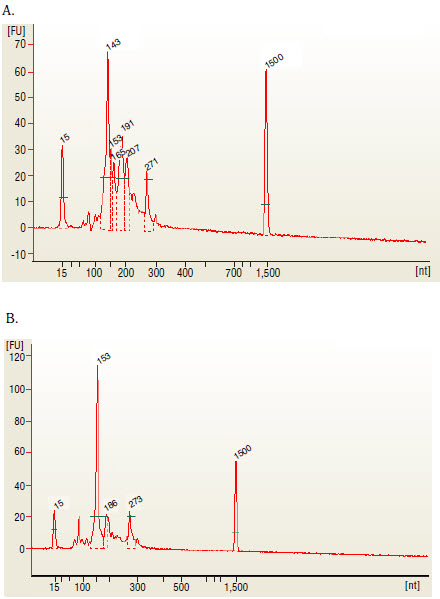
The 143 and 153 bp bands correspond to miRNAs and piRNAs, respectively. The bands on the Bionalyzer electropherograms resolve in sizes ~6-8 nucleotides larger than sizes observed on PAGE gels and can shift from sample to sample due to an incorrect identification of the marker by the bioanalyzer software. miRNA peak should be ~ 143-146 bp.