Overview of High Molecular Weight DNA Extraction using the Monarch® HMW DNA Extraction Kit for Cells & Blood
Script
Extraction of high molecular weight, or HMW DNA, is important for applications that require the use of very long DNA molecules, such as long read sequencing and genome assembly. It is often difficult to extract long DNA that is highly pure and intact, but the Monarch HMW DNA Extraction Kit for Cells and Blood enables this to be done quickly and effectively.
The Monarch method employs a 2-step lysis. First, the cell membrane is lysed with a mild Nuclei Prep Buffer mixed with RNase A. This releases the contents of the cytoplasm but leaves the nuclei intact. The RNA in the cytoplasm is exposed allowing it to be efficiently digested by RNase A. Next, a mix of Nuclei Lysis Buffer and Proteinase K is added to the sample, which lyses the nuclei and releases genomic DNA. Proteinase K digests all of the proteins, including the RNase A enzyme that has finished digesting the RNA. This two-step lysis approach allows the RNA to be digested effectively before the gDNA makes the environment too viscous which would significantly reduce the effectiveness of the RNase A enzyme. When working with blood samples, the leukocytes in the blood, which contain the target genomic DNA, must first be isolated. In mammalian blood, the majority of cells in blood samples are erythrocytes, or red blood cells, which do not contain nuclei, but do contain significant amounts of proteins, mainly hemoglobin. Removing these red blood cells and their proteins is important for a clean blood prep. Red blood cells are first lysed with the RBC Lysis Buffer, and their contents are released into the lysate. Leukocytes remain mostly unharmed by the RBC Lysis Buffer, and they are collected by centrifugation to separate them from the hemoglobin.
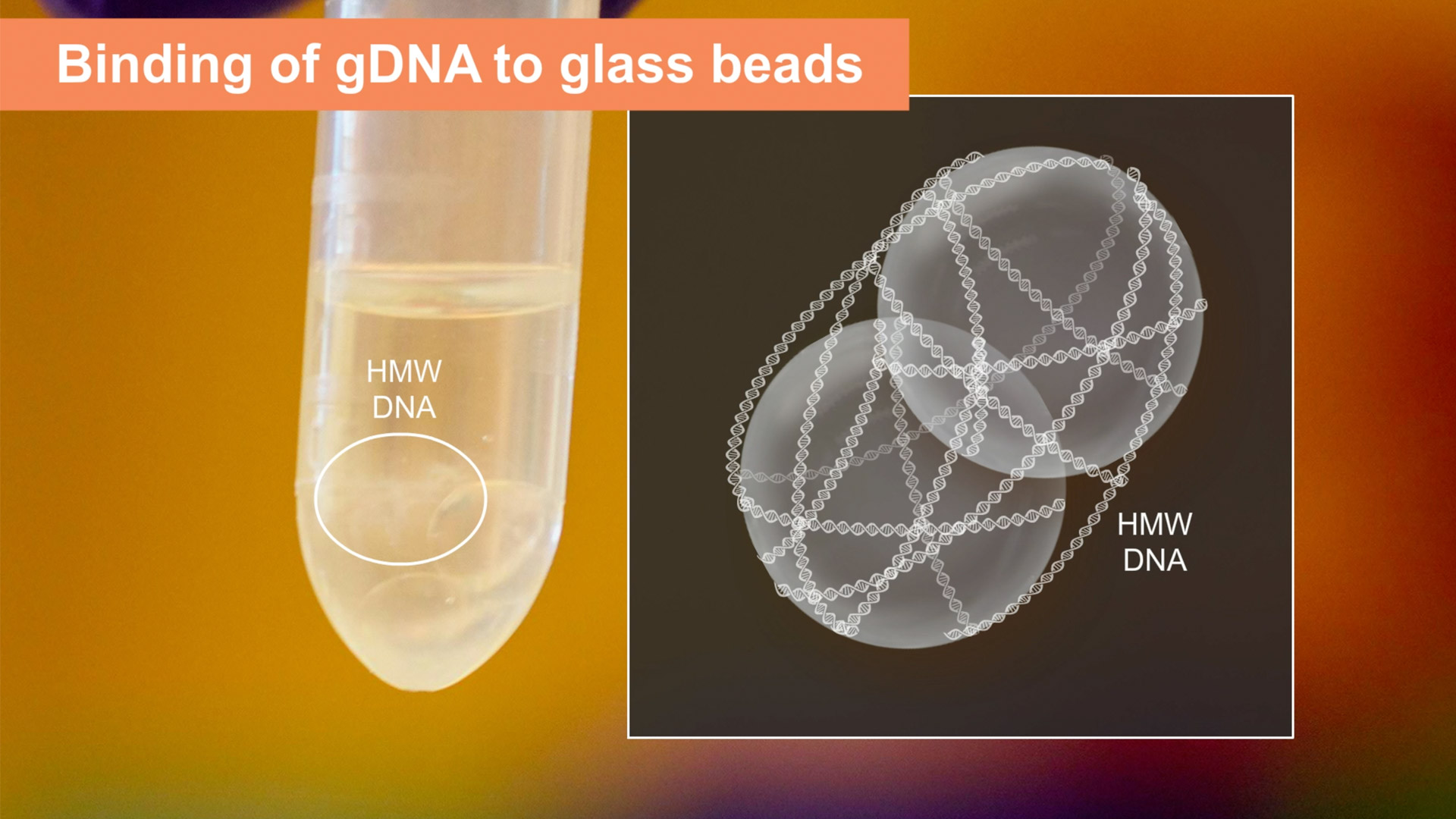
After two wash steps, a clean pellet of leukocytes can be further processed using the cell protocol. During the lysis steps, samples are agitated using a thermal mixer. The speed of agitation determines the fragment length of the DNA. Low or no agitation produces very large DNA, ranging into megabases, while high speeds produce shorter fragments, in the range of 50 to 250 kilobases. Next, alcohol-based precipitation facilitates attachment of the genomic DNA to specialized glass beads. These beads provide a large, smooth surface to collect the precipitated DNA and help prevent shearing. During the binding process, high molecular weight gDNA attaches to the beads and wraps around the large bead surface during inversion. Capturing the DNA on the glass bead surface simplifies the wash and elution steps, eliminating the need for lengthy drying steps or multiple elution steps. After wash buffer is added to the beads, the beads with the DNA bound to them can be poured into the bead retainer which holds onto the beads while a short spin quickly removes all traces of wash buffer and dries the DNA. The beads can then be transferred to a new tube and mixed with elution buffer to begin the elution process. After a few minutes incubation at 56 degrees Celsius, the DNA slides off the smooth surface of the glass beads. The bead retainer is then used again to facilitate the separation of the glass beads from the DNA with a quick an efficient spin, leaving the user with highly pure, high molecular weight DNA. After elution, high molecular weight genomic DNA requires further manipulation before use and measurement.
Eluted DNA will not be uniformly dispersed and often clumps in certain fractions of the solution. Pipetting up and down 5 to 10 times with a wide bore pipet tip helps ensure that any visible DNA clumps are dispersed into the solution. Purified DNA samples can then be stored at 4°C for future use or can be prepared for downstream use as described in the product manual. We invite you to learn more about this kit by visiting www.neb.com/T3050, where you can find a detailed instruction manual and other helpful resources.

