NEB® LAMP Primer Design Tool Tutorial

Script
This video will show you how to design LAMP primers using the New England Biolabs LAMP Primer Design Tool.
To get started, identify the DNA sequence you want to design primers for and select a region of fewer than 2000 bases to input into the tool. This can be loaded as a .txt file or pasted directly into the tool using the “Paste Sequence” feature.
As an example, I’m pasting in a region of <<human BRCA1>> gene sequence.
Once the sequence is pasted in, click the “Process Text” to properly format the sequence text.
If desired, give the sequence target a name in the “Target Name” box but this is not required.
The ‘modify preferences’ box allows for modification of the design parameters. Lengths, Tm, GC content, and many other factors can be adjusted but for any new sequence, we strongly recommend using the default settings for the first attempt, and that’s what we’ll do here.
After the sequence is pasted and processed, click the “Continue” box at the bottom of the first screen.
The next screen displays the target sequence with the ability to specify fixed locations for targeting with one of the LAMP primer regions. This is not common, but if you need a specific sequence to be contained between different primers for a probe or beacon, or a site of variation to be included outside a primer, you can do that here. In most cases, we just click “Generate Primers” at the bottom to start the algorithm identifying suitable LAMP primers.
Click “Generate Loop Primers”. It is possible that no Loop primers can be designed for the candidate Core primer set, in which case a “No Primer Sets Available” box will pop up. If that happens select the next bubble down and try again.
These generated loop primers are not in the “Core” set of 4 LAMP primers from the previous screen and are not required for LAMP, but do provide a significant boost to speed and assay performance and we strongly recommend using them where possible.
In the case that only one primer is identified, we recommend going back to the “Core_Primers” tab or to “select another primer set” and selecting the next set of Core candidates and trying Loop design again.
Choose the top option or one further down the list if you’d like increased GC or another factor.
Select the bubble for the Loop set you like, and then click “Get Selected Primers.”
The final screen will display the full designed primer set, adding in the 2 Loop primers to the previous Core 4 primers on the table.
All relevant information and oligo sequences are on the table, with the Orange sequences representing the oligonucleotides to order from your desired oligonucleotide provider.
Information can be copy/pasted, or you can “Download Primer Set” you’ll be provided with a tab-delimited .txt file directly compatible with batch ordering through the IDT portal.
For any new LAMP assay it is advisable to screen more than one candidate primer set to find the best performance in terms of speed, sensitivity, and non-template amplification.
From the Results tab you can either click “Select More Primer Sets” or the “Core_Primers” tab to go back to the candidate set page. And repeat the selection and Loop primer design steps.
For technical support, visit info@neb.com
Related Videos
-

Behind the Paper: A Simple Isothermal DNA Amplification Method to Screen Black Flies for Onchocerca volvulus Infection -
Behind the Paper: Visual Detection of Isothermal Nucleic Acid Amplification Using pH-sensitive Dyes -
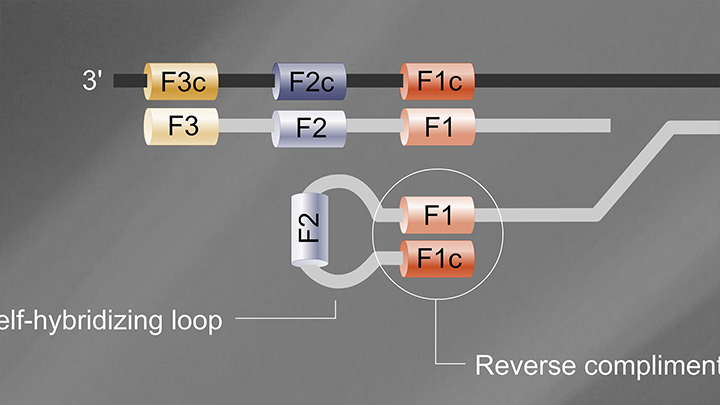
Loop Mediated Isothermal Amplification (LAMP) Tutorial

